Proteomics and Antivenomics of Echis Carinatus Carinatusvenom
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

B1–Proteases As Molecular Targets of Drug Development
Abstracts B1–Proteases as Molecular Targets of Drug Development B1-001 lin release from the beta cells. Furthermore, GLP-1 also stimu- DPP-IV structure and inhibitor design lates beta cell growth and insulin biosynthesis, inhibits glucagon H. B. Rasmussen1, S. Branner1, N. Wagtmann3, J. R. Bjelke1 and secretion, reduces free fatty acids and delays gastric emptying. A. B. Kanstrup2 GLP-1 has therefore been suggested as a potentially new treat- 1Protein Engineering, Novo Nordisk A/S, Bagsvaerd, Denmark, ment for type 2 diabetes. However, GLP-1 is very rapidly degra- 2Medicinal Chemistry, Novo Nordisk A/S, Maaloev, Denmark, ded in the bloodstream by the enzyme dipeptidyl peptidase IV 3Discovery Biology, Novo Nordisk A/S, Maaloev, DENMARK. (DPP-IV; EC 3.4.14.5). A very promising approach to harvest E-mail: [email protected] the beneficial effect of GLP-1 in the treatment of diabetes is to inhibit the DPP-IV enzyme, thereby enhancing the levels of The incretin hormones GLP-1 and GIP are released from the gut endogenously intact circulating GLP-1. The three dimensional during meals, and serve as enhancers of glucose stimulated insu- structure of human DPP-IV in complex with various inhibitors 138 Abstracts creates a better understanding of the specificity and selectivity of drug-like transition-state inhibitors but can be utilized for the this enzyme and allows for further exploration and design of new design of non-transition-state inhibitors that compete for sub- therapeutic inhibitors. The majority of the currently known DPP- strate binding. Besides carrying out proteolytic activity, the IV inhibitors consist of an alpha amino acid pyrrolidine core, to ectodomain of memapsin 2 also interacts with APP leading to which substituents have been added to optimize affinity, potency, the endocytosis of both proteins into the endosomes where APP enzyme selectivity, oral bioavailability, and duration of action. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

Final Program
In Memoriam: Final Program XXV Congress of the International Society on Thrombosis and Haemostasis and 61st Annual SSC Meeting June 20 – 25, 2015 Toronto, Canada www.isth2015.org 1 Final Program Table of Contents 3 Venue and Contacts 5 Invitation and Welcome Message 12 ISTH 2015 Committees 24 Congress Support 25 Sponsors and Exhibitors 27 ISTH Awards 32 ISTH Society Information 37 Program Overview 41 Program Day by Day 55 SSC and Educational Program 83 Master Classes and Career Mentorship Sessions 87 Nurses Forum 93 Scientific Program, Monday, June 22 94 Oral Communications 1 102 Plenary Lecture 103 State of the Art Lectures 105 Oral Communications 2 112 Abstract Symposia 120 Poster Session 189 Scientific Program, Tuesday, June 23 190 Oral Communications 3 198 Plenary Lecture 198 State of the Art Lectures 200 Oral Communications 4 208 Plenary Lecture 209 Abstract Symposia 216 Poster Session 285 Scientific Program, Wednesday, June 24 286 Oral Communications 5 294 Plenary Lecture 294 State of the Art Lectures 296 Oral Communications 6 304 Abstract Symposia 311 Poster Session 381 Scientific Program, Thursday, June 25 382 Oral Communications 7 390 Plenary Lecture 390 Abstract Symposia 397 Highlights of ISTH 399 Exhibition Floor Plan 402 Exhibitor List 405 Congress Information 406 Venue Plan 407 Congress Information 417 Social Program 418 Toronto & Canada Information 421 Transportation in Toronto 423 Future ISTH Meetings and Congresses 2 427 Authors Index 1 Thank You to Everyone Who Supported the Venue and Contacts 2014 World Thrombosis Day -

(12) Patent Application Publication (10) Pub. No.: US 2012/0058468 A1 Mickeown (43) Pub
US 20120058468A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0058468 A1 MickeoWn (43) Pub. Date: Mar. 8, 2012 (54) ADAPTORS FOR NUCLECACID Related U.S. Application Data CONSTRUCTS IN TRANSMEMBRANE (60) Provisional application No. 61/148.737, filed on Jan. SEQUENCING 30, 2009. Publication Classification (75) Inventor: Brian Mckeown, Oxon (GB) (51) Int. Cl. CI2O I/68 (2006.01) (73) Assignee: OXFORD NANOPORE C7H 2L/00 (2006.01) TECHNOLGIES LIMITED, (52) U.S. Cl. ......................................... 435/6.1:536/23.1 Oxford (GB) (57) ABSTRACT (21) Appl. No.: 13/147,159 The invention relates to adaptors for sequencing nucleic acids. The adaptors may be used to generate single stranded constructs of nucleic acid for sequencing purposes. Such (22) PCT Fled: Jan. 29, 2010 constructs may contain both strands from a double stranded deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) (86) PCT NO.: PCT/GB1O/OO160 template. The invention also relates to the constructs gener ated using the adaptors, methods of making the adaptors and S371 (c)(1), constructs, as well as methods of sequencing double stranded (2), (4) Date: Nov. 15, 2011 nucleic acids. Patent Application Publication Mar. 8, 2012 Sheet 1 of 4 US 2012/0058468 A1 Figure 5' 3 Figure 2 Figare 3 Patent Application Publication Mar. 8, 2012 Sheet 2 of 4 US 2012/0058468 A1 Figure 4 End repair Acid adapters ligate adapters Patent Application Publication Mar. 8, 2012 Sheet 3 of 4 US 2012/0058468 A1 Figure 5 Wash away type ifype foducts irrirrosilise Type| capture WashRE products away unbcure Pace É: Wash away unbound rag terts Free told fasgirre?ts aid raiser to festible Figure 6 Beatre Patent Application Publication Mar. -

(5'-Uridylyl)Tyrosine Is the Bond Between the Genome-Linked Protein and the RNA of Poliovirus
Proc. Natl. Acad. Sci. USA Vol. 75, No 10, pp. 4868-4872, October 1978 Biochemistry 04-(5'-Uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus* (iodination/acid and alkali hydrolysis/enzymatic degradation/paper electrophoresis and chromatography) PAUL G. ROTHBERG, TIMOTHY J. R. HARRISt, AKIO NOMOTOt, AND ECKARD WIMMER Department of Microbiology, School of Basic Health Sciences, State University of New York at Stony Brook, Stony Brook, Long Island, New York 11794 Communicated by Seymour S. Cohen, August 3, 1978 ABSTRACT Virion RNA of poliovirus type 1 has been an- MATERIALS AND METHODS alyzed for the linkage between genome-protein VPg and the polyribonucleotide chain. Hydrolysis of the linkage with acid Poliovirus was grown and labeled with phosphorus-32 in HeLa or alkali and enzymatic degradation lead to the conclusion that cell suspension cultures as previously described (9). VPg was the bond is neither a phosphodiester such as nucleotidyl-(P- labeled with [3H]tyrosine as follows: 2.5 X 109 HeLa cells were O-serine (or threonine) nor a phosphoramidate such as washed twice with Earle's saline, suspended in 200 ml of Earle's nucleotidyl4(P-N)amino acid. VPg-RNA can be iodinated by the saline containing 1 mg of actinomycin D, and infected with 50 Bolton and Hunter reagent liodinated 3(4-hydroxyphenyl)pro- plaque-forming units of PV1 per cell. After 25 min at, room pionic acid N-hydroxysuccinimide ester] but not by the chlo- temperature, 200 ml of medium containing 168 ml of Earle's ramine-T or lactoperoxidase procedures, an observation saline, 4 ml of an antibiotic solution (10,000 units of penicillin suggesting that VPg does not contain accessible tyrosine. -

Center for Drug Evaluation and Research Application
CENTER FOR DRUG EVALUATION AND RESEARCH APPLICATION NUMBER: 22-512 PHARMACOLOGY REVIEW(S) Tertiary Pharmacology Review By: Paul C. Brown, Ph.D., ODE Associate Director for Pharmacology and Toxicology OND IO NDA: 22-307 Submission date: 4/19/2010 Drug: Pradaxa™ (Dabigatran etexilate mesylate) Sponsor: Boehringer Ingelheim Pharma GmbH & Co. KG Indication: Prevention of stroke and systemic embolism in patients with atrial fibrillation Reviewing Division: Division of Cardiovascular and Renal Products Comments: The pharm/tox reviewer and supervisor found the nonclinical information submitted for dabigatran to be sufficient to support the proposed use. The reviewer proposes pregnancy category C for the labeling. Dabigatran induced some reproductive toxicity in rats manifest as a decreased number of implantations, decreased number of viable fetuses, increase in the resorption rate, increase in post-implantation loss, and an increase in the number of dead offspring when given at doses of 70 mg/kg (about 2.6 to 3 times the MRHD of 300 mg/day on a mg/m2 basis). Dabigatran also induced some fetal structural variations but did not induce fetal malformations in rats or rabbits. Dabigatran was evaluated for carcinogenicity in 2-year rat and mouse studies. The Executive Carcinogenicity Assessment Committee concluded that these studies were adequate and there were no drug-related tumors in either study. Conclusions: I concur with the Division pharm/tox conclusion that the nonclinical data support approval of this NDA. No additional nonclinical studies are recommended at this time. The proposed Established Pharmacologic Class for dabigatran is "direct thrombin inhibitor". This is appropriate because it is consistent with other moieties of this class. -

Proquest Dissertations
Characterizing the Endoribonuclease Activity of APE1 Wan Cheol Kim BSc, Simon Fraser University, 2007 Thesis Submitted in Partial Fulfillment of The Requirements for the Degree of Master of Science In Mathematical, Computer, and Physical Sciences (Chemistry) The University of Northern British Columbia June 2009 © Wan Cheol Kim, 2009 Library and Archives Bibliotheque et 1*1 Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington OttawaONK1A0N4 Ottawa ON K1A 0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-60814-2 Our file Notre reference ISBN: 978-0-494-60814-2 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library and permettant a la Bibliotheque et Archives Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par I'lnternet, preter, telecommunication or on the Internet, distribuer et vendre des theses partout dans le loan, distribute and sell theses monde, a des fins commerciales ou autres, sur worldwide, for commercial or non support microforme, papier, electronique et/ou commercial purposes, in microform, autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in this et des droits moraux qui protege cette these. Ni thesis. Neither the thesis nor la these ni des extraits substantiels de celle-ci substantial extracts from it may be ne doivent etre imprimes ou autrement printed or otherwise reproduced reproduits sans son autorisation. -

Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases Alan J. Barrett Neil D. Rawlings J. Fred Woessner Editor biographies xxi Contributors xxiii Preface xxxi Introduction ' Abbreviations xxxvii ASPARTIC PEPTIDASES Introduction 1 Aspartic peptidases and their clans 3 2 Catalytic pathway of aspartic peptidases 12 Clan AA Family Al 3 Pepsin A 19 4 Pepsin B 28 5 Chymosin 29 6 Cathepsin E 33 7 Gastricsin 38 8 Cathepsin D 43 9 Napsin A 52 10 Renin 54 11 Mouse submandibular renin 62 12 Memapsin 1 64 13 Memapsin 2 66 14 Plasmepsins 70 15 Plasmepsin II 73 16 Tick heme-binding aspartic proteinase 76 17 Phytepsin 77 18 Nepenthesin 85 19 Saccharopepsin 87 20 Neurosporapepsin 90 21 Acrocylindropepsin 9 1 22 Aspergillopepsin I 92 23 Penicillopepsin 99 24 Endothiapepsin 104 25 Rhizopuspepsin 108 26 Mucorpepsin 11 1 27 Polyporopepsin 113 28 Candidapepsin 115 29 Candiparapsin 120 30 Canditropsin 123 31 Syncephapepsin 125 32 Barrierpepsin 126 33 Yapsin 1 128 34 Yapsin 2 132 35 Yapsin A 133 36 Pregnancy-associated glycoproteins 135 37 Pepsin F 137 38 Rhodotorulapepsin 139 39 Cladosporopepsin 140 40 Pycnoporopepsin 141 Family A2 and others 41 Human immunodeficiency virus 1 retropepsin 144 42 Human immunodeficiency virus 2 retropepsin 154 43 Simian immunodeficiency virus retropepsin 158 44 Equine infectious anemia virus retropepsin 160 45 Rous sarcoma virus retropepsin and avian myeloblastosis virus retropepsin 163 46 Human T-cell leukemia virus type I (HTLV-I) retropepsin 166 47 Bovine leukemia virus retropepsin 169 48 -
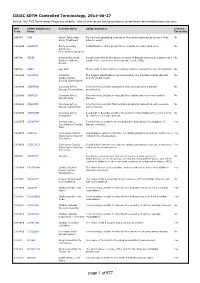
CDISC SDTM Controlled Terminology, 2014-06-27
CDISC SDTM Controlled Terminology, 2014-06-27 Source: NCI EVS Terminology Resources website: http://www.cancer.gov/cancertopics/cancerlibrary/terminologyresources/cdisc NCI CDISC Submission Codelist Name CDISC Definition Codelist Code Value Extensible C66767 ACN Action Taken with Terminology specifying changes to the study treatment as a result of an No Study Treatment adverse event. C101865 ACSPCAT Acute Coronary A classification of the presentation of acute coronary syndrome. No Syndrome Presentation Category C66769 AESEV Severity/Intensity A scale that defines the degree or state of disease existing in a patient as a No Scale for Adverse result of the occurrence of an adverse event. (NCI) Events C66781 AGEU Age Unit Those units of time that are routinely used to express the age of a subject. No C101842 CACRDSC Canadian The anginal classifications as measured by the Canadian Cardiovascular No Cardiovascular Society grading scale. Society Classification C101843 CADPRSN Coronary Artery A terminology codelist associated with coronary artery disease No Disease Presentation presentation. C101849 CADRISK Coronary Artery A terminology codelist to describe the relative risk of coronary artery No Disease Risk disease. C101844 CADSYMP Coronary Artery A terminology codelist that contains symptoms associated with coronary No Disease Symptoms artery disease. C101860 CARTDOM Coronary Artery A codelist to describe whether the posterior descending artery comes from No Dominance the right or left vessel system. C102575 CCINVTYP Contact Case Terminology relevant to the identification and clinical investigation of Yes Investigation Contact disease contacts. Type C101837 CCRCLS Consensus Cardiac Classification system test name for cardiac parameters that are authored or Yes Classification System endorsed by organizations. -

Manual D'estil Per a Les Ciències De Laboratori Clínic
MANUAL D’ESTIL PER A LES CIÈNCIES DE LABORATORI CLÍNIC Segona edició Preparada per: XAVIER FUENTES I ARDERIU JAUME MIRÓ I BALAGUÉ JOAN NICOLAU I COSTA Barcelona, 14 d’octubre de 2011 1 Índex Pròleg Introducció 1 Criteris generals de redacció 1.1 Llenguatge no discriminatori per raó de sexe 1.2 Llenguatge no discriminatori per raó de titulació o d’àmbit professional 1.3 Llenguatge no discriminatori per raó d'ètnia 2 Criteris gramaticals 2.1 Criteris sintàctics 2.1.1 Les conjuncions 2.2 Criteris morfològics 2.2.1 Els articles 2.2.2 Els pronoms 2.2.3 Els noms comuns 2.2.4 Els noms propis 2.2.4.1 Els antropònims 2.2.4.2 Els noms de les espècies biològiques 2.2.4.3 Els topònims 2.2.4.4 Les marques registrades i els noms comercials 2.2.5 Els adjectius 2.2.6 El nombre 2.2.7 El gènere 2.2.8 Els verbs 2.2.8.1 Les formes perifràstiques 2.2.8.2 L’ús dels infinitius ser i ésser 2.2.8.3 Els verbs fer, realitzar i efectuar 2.2.8.4 Les formes i l’ús del gerundi 2.2.8.5 L'ús del verb haver 2.2.8.6 Els verbs haver i caldre 2.2.8.7 La forma es i se davant dels verbs 2.2.9 Els adverbis 2.2.10 Les locucions 2.2.11 Les preposicions 2.2.12 Els prefixos 2.2.13 Els sufixos 2.2.14 Els signes de puntuació i altres signes ortogràfics auxiliars 2.2.14.1 La coma 2.2.14.2 El punt i coma 2.2.14.3 El punt 2.2.14.4 Els dos punts 2.2.14.5 Els punts suspensius 2.2.14.6 El guionet 2.2.14.7 El guió 2.2.14.8 El punt i guió 2.2.14.9 L’apòstrof 2.2.14.10 L’interrogant 2 2.2.14.11 L’exclamació 2.2.14.12 Les cometes 2.2.14.13 Els parèntesis 2.2.14.14 Els claudàtors 2.2.14.15 -

(12) Patent Application Publication (10) Pub. No.: US 2004/0081648A1 Afeyan Et Al
US 2004.008 1648A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2004/0081648A1 Afeyan et al. (43) Pub. Date: Apr. 29, 2004 (54) ADZYMES AND USES THEREOF Publication Classification (76) Inventors: Noubar B. Afeyan, Lexington, MA (51) Int. Cl." ............................. A61K 38/48; C12N 9/64 (US); Frank D. Lee, Chestnut Hill, MA (52) U.S. Cl. ......................................... 424/94.63; 435/226 (US); Gordon G. Wong, Brookline, MA (US); Ruchira Das Gupta, Auburndale, MA (US); Brian Baynes, (57) ABSTRACT Somerville, MA (US) Disclosed is a family of novel protein constructs, useful as Correspondence Address: drugs and for other purposes, termed “adzymes, comprising ROPES & GRAY LLP an address moiety and a catalytic domain. In Some types of disclosed adzymes, the address binds with a binding site on ONE INTERNATIONAL PLACE or in functional proximity to a targeted biomolecule, e.g., an BOSTON, MA 02110-2624 (US) extracellular targeted biomolecule, and is disposed adjacent (21) Appl. No.: 10/650,592 the catalytic domain So that its affinity Serves to confer a new Specificity to the catalytic domain by increasing the effective (22) Filed: Aug. 27, 2003 local concentration of the target in the vicinity of the catalytic domain. The present invention also provides phar Related U.S. Application Data maceutical compositions comprising these adzymes, meth ods of making adzymes, DNA's encoding adzymes or parts (60) Provisional application No. 60/406,517, filed on Aug. thereof, and methods of using adzymes, Such as for treating 27, 2002. Provisional application No. 60/423,754, human Subjects Suffering from a disease, Such as a disease filed on Nov.