Supplementary Data I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human Prion-Like Proteins and Their Relevance in Disease
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Universitat Autònoma de Barcelona Departament de Bioquímica i Biologia Molecular Institut de Biotecnologia i Biomedicina HUMAN PRION-LIKE PROTEINS AND THEIR RELEVANCE IN DISEASE Doctoral thesis presented by Cristina Batlle Carreras for the degree of PhD in Biochemistry, Molecular Biology and Biomedicine from the Universitat Autònoma de Barcelona. The work described herein has been performed in the Department of Biochemistry and Molecular Biology and in the Institute of Biotechnology and Biomedicine, supervised by Prof. Salvador Ventura i Zamora. Cristina Batlle Carreras Prof. Salvador Ventura i Zamora Bellaterra, 2020 Protein Folding and Conformational Diseases Lab. This work was financed with the fellowship “Formación de Profesorado Universitario” by “Ministerio de Ciencia, Innovación y Universidades”. This work is licensed under a Creative Commons Attributions-NonCommercial-ShareAlike 4.0 (CC BY-NC- SA 4.0) International License. The extent of this license does not apply to the copyrighted publications and images reproduced with permission. (CC BY-NC-SA 4.0) Batlle, Cristina: Human prion-like proteins and their relevance in disease. Doctoral Thesis, Universitat Autònoma de Barcelona (2020) English summary ENGLISH SUMMARY Prion-like proteins have attracted significant attention in the last years. -
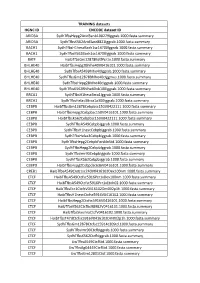
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

SF3B3) and Sin3a Associated Protein 130 (SAP130
cells Communication Ambiguity about Splicing Factor 3b Subunit 3 (SF3B3) and Sin3A Associated Protein 130 (SAP130) Paula I. Metselaar 1,* , Celine Hos 1, Olaf Welting 1, Jos A. Bosch 2,3, Aletta D. Kraneveld 4 , Wouter J. de Jonge 1 and Anje A. Te Velde 1 1 Tytgat Institute for Liver and Intestinal Research, AGEM, Amsterdam UMC, University of Amsterdam, 1105BK Amsterdam, The Netherlands; [email protected] (C.H.); [email protected] (O.W.); [email protected] (W.J.d.J.); [email protected] (A.A.T.V.) 2 Department of Psychology, University of Amsterdam, 1018WS Amsterdam, The Netherlands; [email protected] 3 Department of Medical Psychology, Amsterdam UMC, University of Amsterdam, 1001NK Amsterdam, The Netherlands 4 Division of Pharmacology, Utrecht Institute for Pharmaceutical Sciences, Faculty of Science, Utrecht University, 3584CG Utrecht, The Netherlands; [email protected] * Correspondence: [email protected] Abstract: In 2020, three articles were published on a protein that can activate the immune system by binding to macrophage-inducible C-type lectin receptor (Mincle). In the articles, the protein was referred to as ‘SAP130, a subunit of the histone deacetylase complex.’ However, the Mincle ligand the authors aimed to investigate is splicing factor 3b subunit 3 (SF3B3). This splicing factor is unrelated to SAP130 (Sin3A associated protein 130, a subunit of the histone deacetylase-dependent Sin3A corepressor complex). The conclusions in the three articles were formulated for SF3B3, Citation: Metselaar, P.I.; Hos, C.; while the researchers used qPCR primers and antibodies against SAP130. -

MOZ and MORF, Two Large Mystic Hats in Normal and Cancer Stem Cells
Oncogene (2007) 26, 5408–5419 & 2007 Nature Publishing Group All rights reserved 0950-9232/07 $30.00 www.nature.com/onc REVIEW MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells X-J Yang and M Ullah Molecular Oncology Group, Department of Medicine, McGill University Health Center, Montre´al, Que´bec, Canada Genes of the human monocytic leukemia zinc-finger protein pattern. For cancer biology, it is thus important to MOZ (HUGO symbol, MYST3) and its paralog MORF understand the fundamental mechanisms whereby chro- (MYST4) are rearranged in chromosome translocations matin structure and function are regulated. In the associated withacute myeloid leukemia and/or benign past two decades, our knowledge about regulation in uterine leiomyomata. Both proteins have intrinsic histone this field has exploded. Known regulatory mechanisms acetyltransferase activity and are components of quartet include chromatin assembly, ATP-dependent remodeling, complexes withnoncatalytic subunits containing thebromo- covalent modification, condensin-mediated condensation, domain, plant homeodomain-linked (PHD) finger and replacement with histone variants, and association of proline-tryptophan-tryptophan-proline (PWWP)-containing noncoding RNA (reviewed by Horn and Peterson, 2002; domain, three types of structural modules characteristic of Khorasanizadeh, 2004; Li et al., 2007). Covalent chromatin regulators. Although leukemia-derived fusion pro- modification can occur at both the DNA and histone teins suchas MOZ-TIF2 promote self-renewal of leukemic levels. With histones, modifications include acetylation, stem cells, recent studies indicate that murine MOZ and phosphorylation, methylation, ubiquitination, and many MORF are important for proper development of hema- others (for reviews, see Spencer and Davie, 1999; Strahl topoietic and neurogenic progenitors, respectively, thereby and Allis, 2000; Berger, 2002; Jason et al., 2002; highlighting the importance of epigenetic integrity in Kouzarides, 2007). -

Genome-Wide Inference of Natural Selection on Human Transcription Factor Binding Sites
ANALYSIS Genome-wide inference of natural selection on human transcription factor binding sites Leonardo Arbiza1, Ilan Gronau1, Bulent A Aksoy2, Melissa J Hubisz1, Brad Gulko3, Alon Keinan1–3 & Adam Siepel1–3 For decades, it has been hypothesized that gene regulation persistence in humans7,8. In addition, some genome-wide analyses has had a central role in human evolution, yet much remains have found bulk statistical evidence of natural selection in noncoding unknown about the genome-wide impact of regulatory regions near genes, presumably due to cis-regulatory elements9–12. mutations. Here we use whole-genome sequences and genome- Nevertheless, evidence in support of the overall prominence of wide chromatin immunoprecipitation and sequencing data to cis-regulatory mutations in evolutionary adaptation remains largely demonstrate that natural selection has profoundly influenced anecdotal and indirect, and there is continuing controversy about the human transcription factor binding sites since the divergence relative roles of regulatory and protein-coding sequences in evolu- of humans from chimpanzees 4–6 million years ago. Our tion8. Large-scale genomic studies of the evolution of transcription analysis uses a new probabilistic method, called INSIGHT, for factor binding sites have the potential to advance this debate, but a measuring the influence of selection on collections of short, major limitation of such studies so far has been a lack of precisely interspersed noncoding elements. We find that, on average, annotated binding sites across the genome. The analysis of con- transcription factor binding sites have experienced somewhat served noncoding sequences and/or promoter regions rather than weaker selection than protein-coding genes. -

Mediator of DNA Damage Checkpoint 1 (MDC1) Is a Novel Estrogen Receptor Co-Regulator in Invasive 6 Lobular Carcinoma of the Breast 7 8 Evelyn K
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.16.423142; this version posted December 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Running Title: MDC1 co-regulates ER in ILC 2 3 Research article 4 5 Mediator of DNA damage checkpoint 1 (MDC1) is a novel estrogen receptor co-regulator in invasive 6 lobular carcinoma of the breast 7 8 Evelyn K. Bordeaux1+, Joseph L. Sottnik1+, Sanjana Mehrotra1, Sarah E. Ferrara2, Andrew E. Goodspeed2,3, James 9 C. Costello2,3, Matthew J. Sikora1 10 11 +EKB and JLS contributed equally to this project. 12 13 Affiliations 14 1Dept. of Pathology, University of Colorado Anschutz Medical Campus 15 2Biostatistics and Bioinformatics Shared Resource, University of Colorado Comprehensive Cancer Center 16 3Dept. of Pharmacology, University of Colorado Anschutz Medical Campus 17 18 Corresponding author 19 Matthew J. Sikora, PhD.; Mail Stop 8104, Research Complex 1 South, Room 5117, 12801 E. 17th Ave.; Aurora, 20 CO 80045. Tel: (303)724-4301; Fax: (303)724-3712; email: [email protected]. Twitter: 21 @mjsikora 22 23 Authors' contributions 24 MJS conceived of the project. MJS, EKB, and JLS designed and performed experiments. JLS developed models 25 for the project. EKB, JLS, SM, and AEG contributed to data analysis and interpretation. SEF, AEG, and JCC 26 developed and performed informatics analyses. MJS wrote the draft manuscript; all authors read and revised the 27 manuscript and have read and approved of this version of the manuscript. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genome-Wide Profiling of Active Enhancers in Colorectal Cancer
Genome-wide proling of active enhancers in colorectal cancer Min Wu ( [email protected] ) Wuhan University https://orcid.org/0000-0003-1372-4764 Qinglan Li Wuhan University Xiang Lin Wuhan University Ya-Li Yu Zhongnan Hospital, Wuhan University Lin Chen Wuhan University Qi-Xin Hu Wuhan University Meng Chen Zhongnan Hospital, Wuhan University Nan Cao Zhongnan Hospital, Wuhan University Chen Zhao Wuhan University Chen-Yu Wang Wuhan University Cheng-Wei Huang Wuhan University Lian-Yun Li Wuhan University Mei Ye Zhongnan Hospital, Wuhan University https://orcid.org/0000-0002-9393-3680 Article Keywords: Colorectal cancer, H3K27ac, Epigenetics, Enhancer, Transcription factors Posted Date: December 10th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-119156/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Genome-wide profiling of active enhancers in colorectal cancer Qing-Lan Li1, #, Xiang Lin1, #, Ya-Li Yu2, #, Lin Chen1, #, Qi-Xin Hu1, Meng Chen2, Nan Cao2, Chen Zhao1, Chen-Yu Wang1, Cheng-Wei Huang1, Lian-Yun Li1, Mei Ye2,*, Min Wu1,* 1 Frontier Science Center for Immunology and Metabolism, Hubei Key Laboratory of Cell Homeostasis, Hubei Key Laboratory of Developmentally Originated Disease, Hubei Key Laboratory of Intestinal and Colorectal Diseases, College of Life Sciences, Wuhan University, Wuhan, Hubei 430072, China 2Division of Gastroenterology, Department of Geriatrics, Hubei Clinical Centre & Key Laboratory of Intestinal and Colorectal Diseases, Zhongnan Hospital, Wuhan University, Wuhan, Hubei 430072, China #Equal contribution to the study. Contact information *Correspondence should be addressed to Dr. Min Wu, Email: [email protected], Tel: 86-27-68756620, or Dr. -

Functional Characterization of the New 8Q21 Asthma Risk Locus
Functional characterization of the new 8q21 Asthma risk locus Cristina M T Vicente B.Sc, M.Sc A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2017 Faculty of Medicine Abstract Genome wide association studies (GWAS) provide a powerful tool to identify genetic variants associated with asthma risk. However, the target genes for many allergy risk variants discovered to date are unknown. In a recent GWAS, Ferreira et al. identified a new association between asthma risk and common variants located on chromosome 8q21. The overarching aim of this thesis was to elucidate the biological mechanisms underlying this association. Specifically, the goals of this study were to identify the gene(s) underlying the observed association and to study their contribution to asthma pathophysiology. Using genetic data from the 1000 Genomes Project, we first identified 118 variants in linkage disequilibrium (LD; r2>0.6) with the sentinel allergy risk SNP (rs7009110) on chromosome 8q21. Of these, 35 were found to overlap one of four Putative Regulatory Elements (PREs) identified in this region in a lymphoblastoid cell line (LCL), based on epigenetic marks measured by the ENCODE project. Results from analysis of gene expression data generated for LCLs (n=373) by the Geuvadis consortium indicated that rs7009110 is associated with the expression of only one nearby gene: PAG1 - located 732 kb away. PAG1 encodes a transmembrane adaptor protein localized to lipid rafts, which is highly expressed in immune cells. Results from chromosome conformation capture (3C) experiments showed that PREs in the region of association physically interacted with the promoter of PAG1. -
![Downloaded from JASPAR (JASPAR ID: MA0527.1) Across All Identified Peaks Using the Matrix-Scan Module from RSAT [20]](https://docslib.b-cdn.net/cover/5028/downloaded-from-jaspar-jaspar-id-ma0527-1-across-all-identified-peaks-using-the-matrix-scan-module-from-rsat-20-355028.webp)
Downloaded from JASPAR (JASPAR ID: MA0527.1) Across All Identified Peaks Using the Matrix-Scan Module from RSAT [20]
bioRxiv preprint doi: https://doi.org/10.1101/585653; this version posted March 24, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. ZBTB33 (Kaiso) methylated binding sites are associated with primed heterochromatin Quy Xiao Xuan Lin1, Khadija Rebbani1, Sudhakar Jha1,2, Touati Benoukraf1,3* 1Cancer Science Institute of Singapore, National University of Singapore, Singapore, Singapore 2Department of Biochemistry, National University of Singapore, Singapore, Singapore 3Discipline of Genetics, Faculty of Medicine, Memorial University of Newfoundland, St. John’s, NL, Canada *Correspondence to: Touati Benoukraf, Ph.D. Faculty of Medicine, Discipline of Genetics Cancer Science Institute of Singapore Craig L. Dobbin Genetics Research Centre National University of Singapore Room 5M317 Centre for Translational Medicine, Memorial University of Newfoundland 14 Medical Drive, #12-01 St. John's, NL A1B 3V6 Singapore 117599 Canada Phone: +1 (709) 864-6671 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/585653; this version posted March 24, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Background: ZBTB33, also known as Kaiso, is a member of the zinc finger and BTB/POZ family. In contrast to many transcription factors, ZBTB33 has the ability to bind both a sequence-specific consensus and methylated DNA. -

The Basic Region and Leucine Zipper Transcription Factor Mafk Is a New Nerve Growth Factor-Responsive Immediate Early Gene That Regulates Neurite Outgrowth
The Journal of Neuroscience, October 15, 2002, 22(20):8971–8980 The Basic Region and Leucine Zipper Transcription Factor MafK Is a New Nerve Growth Factor-Responsive Immediate Early Gene That Regulates Neurite Outgrowth Be´ ata To¨ro¨ csik, James M. Angelastro, and Lloyd A. Greene Department of Pathology and Center for Neurobiology and Behavior, Columbia University College of Physicians and Surgeons, New York, New York 10032 We used serial analysis of gene expression to identify new mediated by an atypical isoform of PKC but not by mitogen- NGF-responsive immediate early genes (IEGs) with potential activated kinase kinase, phospholipase C␥, or phosphoinositide roles in neuronal differentiation. Among those identified was 3Ј-kinase. Interference with MafK expression or activity by small MafK, a small Maf family basic region and leucine zipper tran- interfering RNA and dominant negative strategies, respectively, scriptional repressor and coactivator expressed in immature suppresses NGF-promoted outgrowth and maintenance of neu- neurons. NGF treatment elevates the levels of both MafK tran- rites by PC12 cells and neurite outgrowth by immature telence- scripts and protein. In contrast, there is no effect on expression phalic neurons. Our findings support a role for MafK as a novel of the closely related MafG. Unlike many other NGF-responsive regulator of neuronal differentiation. IEGs, MafK regulation shows selectivity and is unresponsive to epidermal growth factor, depolarization, or cAMP derivatives. Key words: MafK; NGF; immediate early -

Hepatitis C Virus As a Unique Human Model Disease to Define
viruses Review Hepatitis C Virus as a Unique Human Model Disease to Define Differences in the Transcriptional Landscape of T Cells in Acute versus Chronic Infection David Wolski and Georg M. Lauer * Liver Center at the Gastrointestinal Unit, Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA * Correspondence: [email protected]; Tel.: +1-617-724-7515 Received: 27 June 2019; Accepted: 23 July 2019; Published: 26 July 2019 Abstract: The hepatitis C virus is unique among chronic viral infections in that an acute outcome with complete viral elimination is observed in a minority of infected patients. This unique feature allows direct comparison of successful immune responses with those that fail in the setting of the same human infection. Here we review how this scenario can be used to achieve better understanding of transcriptional regulation of T-cell differentiation. Specifically, we discuss results from a study comparing transcriptional profiles of hepatitis C virus (HCV)-specific CD8 T-cells during early HCV infection between patients that do and do not control and eliminate HCV. Identification of early gene expression differences in key T-cell differentiation molecules as well as clearly distinct transcriptional networks related to cell metabolism and nucleosomal regulation reveal novel insights into the development of exhausted and memory T-cells. With additional transcriptional studies of HCV-specific CD4 and CD8 T-cells in different stages of infection currently underway, we expect HCV infection to become a valuable model disease to study human immunity to viruses. Keywords: viral hepatitis; hepatitis C virus; T cells; transcriptional regulation; transcription factors; metabolism; nucleosome 1.