Characterisation of Fungi of Stored Common Bean Cultivars Grown in Menoua Division, Cameroon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Page 1 C H a D N I G E R N I G E R I a G a B O N CENTRAL AFRICAN
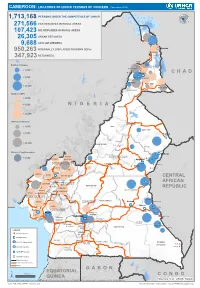
CAMEROON: LOCATIONS OF UNHCR PERSONS OF CONCERN (November 2019) 1,713,168 PERSONS UNDER THE COMPETENCENIGER OF UNHCR 271,566 CAR REFUGEES IN RURAL AREAS 107,423 NIG REFUGEES IN RURAL AREAS 26,305 URBAN REFUGEES 9,688 ASYLUM SEEKERS 950,263 INTERNALLY DISPLACED PERSONS (IDPs) Kousseri LOGONE 347,923 RETURNEES ET CHARI Waza Limani Magdeme Number of refugees EXTRÊME-NORD MAYO SAVA < 3,000 Mora Mokolo Maroua CHAD > 5,000 Minawao DIAMARÉ MAYO TSANAGA MAYO KANI > 20,000 MAYO DANAY MAYO LOUTI Number of IDPs < 2,000 > 5,000 NIGERIA BÉNOUÉ > 20,000 Number of returnees NORD < 2,000 FARO MAYO REY > 5,000 Touboro > 20,000 FARO ET DÉO Beke chantier Ndip Beka VINA Number of asylum seekers Djohong DONGA < 5,000 ADAMAOUA Borgop MENCHUM MANTUNG Meiganga Ngam NORD-OUEST MAYO BANYO DJEREM Alhamdou MBÉRÉ BOYO Gbatoua BUI Kounde MEZAM MANYU MOMO NGO KETUNJIA CENTRAL Bamenda NOUN BAMBOUTOS AFRICAN LEBIALEM OUEST Gado Badzere MIFI MBAM ET KIM MENOUA KOUNG KHI REPUBLIC LOM ET DJEREM KOUPÉ HAUTS PLATEAUX NDIAN MANENGOUBA HAUT NKAM SUD-OUEST NDÉ Timangolo MOUNGO MBAM ET HAUTE SANAGA MEME Bertoua Mbombe Pana INOUBOU CENTRE Batouri NKAM Sandji Mbile Buéa LITTORAL KADEY Douala LEKIÉ MEFOU ET Lolo FAKO AFAMBA YAOUNDE Mbombate Yola SANAGA WOURI NYONG ET MARITIME MFOUMOU MFOUNDI NYONG EST Ngarissingo ET KÉLLÉ MEFOU ET HAUT NYONG AKONO Mboy LEGEND Refugee location NYONG ET SO’O Refugee Camp OCÉAN MVILA UNHCR Representation DJA ET LOBO BOUMBA Bela SUD ET NGOKO Libongo UNHCR Sub-Office VALLÉE DU NTEM UNHCR Field Office UNHCR Field Unit Region boundary Departement boundary Roads GABON EQUATORIAL 100 Km CONGO ± GUINEA The boundaries and names shown and the designations used on this map do not imply official endorsement or acceptance by the United Nations Sources: Esri, USGS, NOAA Source: IOM, OCHA, UNHCR – Novembre 2019 Pour plus d’information, veuillez contacter Jean Luc KRAMO ([email protected]). -

Cameroon Watershed
Journal(of(Materials(and(( J. Mater. Environ. Sci., 2018, Volume 9, Issue 11, Page 3094-2104 Environmental(Sciences( ISSN(:(2028;2508( CODEN(:(JMESCN( http://www.jmaterenvironsci.com! Copyright(©(2018,( University(of(Mohammed(Premier(((((( (Oujda(Morocco( A typological study of the physicochemical quality of water in the Menoua- Cameroon watershed Santsa Nguefack Charles Vital1*, Ndjouenkeu Robert1, Ngassoum Martin Benoit2 , 1Laboratory of Food Physicochemistry, Department of Food Science and Nutrition National Advanced School of Agro- Industrial Sciences, University of Ngaoundéré, PO BOX 455,Ngaoundéré, Cameroon. 2Department of Industrial Chemistry and Environment, National Advanced School of Agro-Industrial Sciences, University of Ngaoundéré, PO BOX 455, Ngaoundéré, Cameroon. Received 29 Oct 2017 Abstract Revised 25 Dec 2018 Cameroon, a country in Central Africa with an outlet to the Atlantic Ocean in the Gulf Accepted 28 Dec 2018 of Guinea, holds enormous potentialities of water resources. These resources used for the supply of water for human consumption, etc., would be polluted by the Keywords intensification of anthropogenic activities, of which the most important recorded in the !!physicochemical, Menoua watershedof West Cameroon are agriculture and breeding. The objective of this !!pollution, work was to study the spatiotemporal parameters of the physicochemical quality !!Menoua watershed, parameters (temperature, pH, electrical conductivity, Turbidity, Nitrates, Ortho !!Anthropogenic Phosphates, Sulphates, Chloride, Potassium and Calcium) the Menoua activities, watershedthrough a main Principal Component Analysis (ACP). The results obtained showed that the pH, turbidity and orthophosphate have levels which are in contradiction !!Water for human consumption. with WHO standards. The physicochemical parameters of this water showed significant spatial and seasonal variations, the causes of which probably related to the human activities generating pollutants. -

An Inventory of Short Horn Grasshoppers in the Menoua Division, West Region of Cameroon
AGRICULTURE AND BIOLOGY JOURNAL OF NORTH AMERICA ISSN Print: 2151-7517, ISSN Online: 2151-7525, doi:10.5251/abjna.2013.4.3.291.299 © 2013, ScienceHuβ, http://www.scihub.org/ABJNA An inventory of short horn grasshoppers in the Menoua Division, West Region of Cameroon Seino RA1, Dongmo TI1, Ghogomu RT2, Kekeunou S3, Chifon RN1, Manjeli Y4 1Laboratory of Applied Ecology (LABEA), Department of Animal Biology, Faculty of Science, University of Dschang, P.O. Box 353 Dschang, Cameroon, 2Department of Plant Protection, Faculty of Agriculture and Agronomic Sciences (FASA), University of Dschang, P.O. Box 222, Dschang, Cameroon. 3 Département de Biologie et Physiologie Animale, Faculté des Sciences, Université de Yaoundé 1, Cameroun 4 Department of Biotechnology and Animal Production, Faculty of Agriculture and Agronomic Sciences (FASA), University of Dschang, P.O. Box 222, Dschang, Cameroon. ABSTRACT The present study was carried out as a first documentation of short horn grasshoppers in the Menoua Division of Cameroon. A total of 1587 specimens were collected from six sites i.e. Dschang (265), Fokoue (253), Fongo – Tongo (267), Nkong – Ni (271), Penka Michel (268) and Santchou (263). Identification of these grasshoppers showed 28 species that included 22 Acrididae and 6 Pyrgomorphidae. The Acrididae belonged to 8 subfamilies (Acridinae, Catantopinae, Cyrtacanthacridinae, Eyprepocnemidinae, Oedipodinae, Oxyinae, Spathosterninae and Tropidopolinae) while the Pyrgomorphidae belonged to only one subfamily (Pyrgomorphinae). The Catantopinae (Acrididae) showed the highest number of species while Oxyinae, Spathosterninae and Tropidopolinae showed only one species each. Ten Acrididae species (Acanthacris ruficornis, Anacatantops sp, Catantops melanostictus, Coryphosima stenoptera, Cyrtacanthacris aeruginosa, Eyprepocnemis noxia, Gastrimargus africanus, Heteropternis sp, Ornithacris turbida, and Trilophidia conturbata ) and one Pyrgomorphidae (Zonocerus variegatus) were collected in all the six sites. -

N I G E R I a C H a D Central African Republic Congo
CAMEROON: LOCATIONS OF UNHCR PERSONS OF CONCERN (September 2020) ! PERSONNES RELEVANT DE Maïné-Soroa !Magaria LA COMPETENCE DU HCR (POCs) Geidam 1,951,731 Gashua ! ! CAR REFUGEES ING CurAi MEROON 306,113 ! LOGONE NIG REFUGEES IN CAMEROON ET CHARI !Hadejia 116,409 Jakusko ! U R B A N R E F U G E E S (CENTRAL AFRICAN REPUBLIC AND 27,173 NIGERIAN REFUGEE LIVING IN URBAN AREA ARE INCLUDED) Kousseri N'Djamena !Kano ASYLUM SEEKERS 9,332 Damaturu Maiduguri Potiskum 1,032,942 INTERNALLY DISPLACED PERSO! NS (IDPs) * RETURNEES * Waza 484,036 Waza Limani Magdeme Number of refugees MAYO SAVA Mora ! < 10,000 EXTRÊME-NORD Mokolo DIAMARÉ Biu < 50,000 ! Maroua ! Minawao MAYO Bauchi TSANAGA Yagoua ! Gom! be Mubi ! MAYO KANI !Deba MAYO DANAY < 75000 Kaele MAYO LOUTI !Jos Guider Number! of IDPs N I G E R I A Lafia !Ləre ! < 10,000 ! Yola < 50,000 ! BÉNOUÉ C H A D Jalingo > 75000 ! NORD Moundou Number of returnees ! !Lafia Poli Tchollire < 10,000 ! FARO MAYO REY < 50,000 Wukari ! ! Touboro !Makurdi Beke Chantier > 75000 FARO ET DÉO Tingere ! Beka Paoua Number of asylum seekers Ndip VINA < 10,000 Bocaranga ! ! Borgop Djohong Banyo ADAMAOUA Kounde NORD-OUEST Nkambe Ngam MENCHUM DJEREM Meiganga DONGA MANTUNG MAYO BANYO Tibati Gbatoua Wum BOYO MBÉRÉ Alhamdou !Bozoum Fundong Kumbo BUI CENTRAL Mbengwi MEZAM Ndop MOMO AFRICAN NGO Bamenda KETUNJIA OUEST MANYU Foumban REPUBLBICaoro BAMBOUTOS ! LEBIALEM Gado Mbouda NOUN Yoko Mamfe Dschang MIFI Bandjoun MBAM ET KIM LOM ET DJEREM Baham MENOUA KOUNG KHI KOUPÉ Bafang MANENGOUBA Bangangte Bangem HAUT NKAM Calabar NDÉ SUD-OUEST -

Shelter Cluster Cameroon – Meeting Minutes Meeting: Shelter Cluster
Shelter Cluster Cameroon ShelterCluster.org Coordinating Humanitarian Shelter Yaounde 30 October 2020 Shelter Cluster Cameroon – Meeting Minutes Meeting: Shelter Cluster Date: 29.10.2020 Time: 15:00 Meeting Facilitator: Medar Mitima Kajemba Location: Yaounde Minutes Prepared By: Medar Mitima Kajemba Location: UNHCR Office Agenda • Introduction and round table • Agreement on frequency of Yaounde shelter cluster meeting. • Shelter-NFI activities-presentation-Who is doing What and Where. • Current Shelter-NFI needs(gaps) in the NW/SW regions. • Recommendation • AOB Introduction and round table Everyone introduced themselves by name, organization and position in the organization, which allowed participants to get to know each other Agreement on frequency of Yaounde shelter cluster meeting. The schedule of meetings will be set after consultation with the field clusters to ensure that inputs from the field will be available before each meeting in Yaounde Shelter-NFI activities-presentation-Who is doing What and Where Catholic Relief Services (CRS): CRS's shelter and resettlement program is part of its RRF (Rapid Response Fund) program, which has just been launched in 4 African countries including Burkina Faso, Mali, Niger and Cameroon. The West Africa RRF mechanism is designed to meet unanticipated needs as they emerge, ensuring rapid, short-term, life-saving assistance through Water Sanitation and Hygiene (WASH), Multi-Purpose Cash (MPCA), and Shelter and Settlement (S&S) interventions. Through organizational and emergency response capacity building, the project will emphasize the use of local partner networks to deepen the potential for sustainability and transition. In Cameroon, the project will cover Far North, East, North West and South West regions. -

Infected Areas As on 9 June 1988 — Zones Infectées Au 9 Juin 1988 for Criteria Used in Compiling This List, See No
Wklv Eptdem Rec : No. 24 - 10 June 1988 - 182 - Relevé àpidém bebd. : N° 24 - 10 juin 1988 (i) 423 notifications saved in the 15 years following 100 000 i) 423 notifications évitées au cours des 15 années suivant les immunizations in the years 1972-1976; 100 000 vaccinations administrées de 1972 à 1976; (Ü) 212 notifications saved in the 15 years following 100 000 ii) 212 .notifications .évitées au cours des 15 années suivant les immunizations in the years 1977-1981 ; 100000 vaccinations administrées de 1977 à 1981; (iii) 100 notifications saved in the 15 years following 100 000 iii) 100 notifications'évitées au cours des 15 années suivant les . immunizations in the years 1982-1986. 100 000 vaccinations administrées de 1982 à 1986. ' Approximately 65 000 BCG immunizations are given annually Quelque 65 000 vaccinations par le BCG sont administrées chaque in Scotland, therefore the saving per year is estimated at 65 cases année en Ecosse; on estime donc à 65 le nombre des cas évités chaque in the 15-29 year age group. année dans le groupe d’âge 15-29 ans. To stop using BCG would mean an increase in disease among Ne plus utiliser le BCG entraînerait Une progression de la maladie members of the 15-29 year age group, local outbreaks would be chez les 15-29 ans, un risque accru de poussées locales du fait de la perte expected if herd resistance is lost and the individual would be at de résistance collective et une augmentation du risque individuel pour increased risk when moving from an area of low infectivity in quiconque pénétrerait dans une zone de haute infectivité, au Royaume- the United Kingdom to an area of high infectivity either in the Uni ou à l’étranger, en provenance d’une zone de faible infectivité au United Kingdom or abroad. -

Pdf | 300.72 Kb
Report Multi-Sector Rapid Assessment in the West and Littoral Regions Format Cameroon, 25-29 September 2018 1. GENERAL OVERVIEW a) Background What? The humanitarian crisis affecting the North-West and the South-West Regions has a growing impact in the bordering regions of West and Littoral. Since April 2018, there has been a proliferation of non-state armed groups (NSAG) and intensification of confrontations between NSAG and the state armed forces. As of 1st October, an estimated 350,000 people are displaced 246,000 in the South-West and 104,000 in the North-West; with a potential increment due to escalation in hostilities. Why? An increasing number of families are leaving these regions to take refuge in Littoral and the West Regions following disruption of livelihoods and agricultural activities. Children are particularly affected due to destruction or closure of schools and the “No School” policy ordered by NSAG since 2016. The situation has considerably evolved in the past three months because of: i) the anticipated security flashpoints (the start of the school year, the “October 1st anniversary” and the elections); ii) the increasing restriction of movement (curfew extended in the North-West, “No Movement Policy” issued by non-state actors; and iii) increase in both official and informal checkpoints. Consequently, there has been a major increase in the number of people leaving the two regions to seek safety and/or to access economic and educational opportunities. Preliminary findings indicate that IDPs are facing similar difficulties and humanitarian needs than the one reported in the North-West and the South-West regions following the multisectoral needs assessment done in March 2018. -

Cameroon Developing Subnational Estimates of Hiv Prevalence and the Number of People
UNAIDS 2014 | REFERENCE CAMEROON DEVELOPING SUBNATIONAL ESTIMATES OF HIV PREVALENCE AND THE NUMBER OF PEOPLE LIVING WITH HIV UNAIDS / JC2665E (English original, September 2014) Copyright © 2014. Joint United Nations Programme on HIV/AIDS (UNAIDS). All rights reserved. Publications produced by UNAIDS can be obtained from the UNAIDS Information Production Unit. Reproduction of graphs, charts, maps and partial text is granted for educational, not-for-profit and commercial purposes as long as proper credit is granted to UNAIDS: UNAIDS + year. For photos, credit must appear as: UNAIDS/name of photographer + year. Reproduction permission or translation-related requests—whether for sale or for non-commercial distribution—should be addressed to the Information Production Unit by e-mail at: [email protected]. The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of UNAIDS concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. UNAIDS does not warrant that the information published in this publication is complete and correct and shall not be liable for any damages incurred as a result of its use. METHODOLOGY NOTE Developing subnational estimates of HIV prevalence and the number of people living with HIV from survey data Introduction prevR Significant geographic variation in HIV Applying the prevR method to generate maps incidence and prevalence, as well as of estimates of the number of people living programme implementation, has been with HIV (aged 15–49 and 15 and older) and observed between and within countries. -

Dynamiques Territoriales De La Production Maraichère Dans Les Hauts
Dynamiques territoriales de la production maraichère dans les Hauts Plateaux de l’Ouest Cameroun : cas du département de la Menoua Wuld Daniel Paddy MVENG1* ; Abdoulay NSANGOU NJANKOUO2 ; Fabrice Parfait AZEBAZE KENFACK3 ; Marcien KUETE FOGANG4 ; Laure Lysette CHIMI NKOMBO5 ; Irène LAKEU MELI6 1- Economiste du territoire, chercheur IRAD (Institut de Recherche Agricole pour le Développement) 2- Ingénieur Agro-socio-économiste, chercheur IRAD 3- Socio-Economiste, chercheur IRAD 4- Géographe-cartographe/ agroforestier, chercheur IRAD 5- Environnementaliste, chercheur IRAD 6- Environnementaliste, chercheur IRAD Résumé: Le maraîchage s'est développé rapidement au cours des dernières décennies au Cameroun. Certaines zones agroécologiques comme les Hauts Plateaux de l'Ouest, sont devenues des bassins maraîchers importants. Cet article examine les dynamiques récentes de la production maraîchère dans cette zone à travers le département de la Menoua. L'objectif est non seulement de déterminer le poids des principales cultures dans les arrondissements du département, mais surtout de l'analyser afin de mettre en évidence leurs dynamiques territoriales. Pour ce faire, treize cultures maraîchères ont été échantillonnées sur la base des données fournies par la Délégation Départementale de l'Agriculture. Leur traitement visait à mesurer l'indice de spécialisation territoriale ( ) sur ces cultures. Globalement, il ressort que la production maraichère du département푎 푆푝é sur la période 2010-2020 présente des situations territoriales contrastées. Il est aussi observé que les arrondissements se diversifient plus qu’ils ne se spécialisent. Si la tendance à la diversification concerne Dschang, Fongo-Tongo, Knong-Gni et Penka-Michel, la tendance à la spécialisation ne concerne que Fokoué et Santchou. Des facteurs économiques et territoriaux justifient ces 59 résultats. -

CAMEROON: Administrative Map with Locations of Persons of Concerns October 2016 NIGER Lake Chad
CAMEROON: Administrative map with locations of Persons of Concerns October 2016 NIGER Lake Chad 259,145 CAR REFUGEES Logone-Et-Chari 73,747 NIG REFUGEES Kousseri 19,802 URBAN REFUGEES Waza IDPs Limani 192,912 Magdeme Mora Mayo-Sava IDP RETURNEES Diamare 32,023 Mokolo REP. OF Gawar EXTREME-NORD Minawao Maroua CHAD Mayo-Tsanaga Mayo-Kani Mayo-Danay Mayo-Louti NIGERIA number of refugees in camp Benoue >5000 >15000 NORD Faro >20,000 Mayo-Rey number of refugees out of camp >3000 >5000 Faro-et-Deo Beke chantier >20,000 Vina Ndip Beka Borgop Nyambaka number of urban refugees ADAMAOUA Djohong Ngam Gbata Alhamdou Menchum Donga-Mantung >5000 Meiganga Mayo-Banyo Djerem Mbere Kounde Gadi Akwaya NORD-OUEST Gbatoua Boyo Bui Foulbe <10,000 Mbale Momo Mezam number of IDPs Ngo-ketunjia Manyu Gado Bamboutos Badzere <2000 Lebialem Noun Mifi Sodenou >5000 Menoua OUEST Mbam-et-Kim CENTRAL Hauts-Plateaux Lom-Et-Djerem Kupe-Manenguba Koung-Khi AFRICAN >20,000 Haut-Nkam SUD-OUEST Nde REPUBLIC Ndian Haute-Sanaga Mbam-et-Inoubou Moinam Meme CENTRE Timangolo Bertoua Bombe Sandji1 Nkam Batouri Pana Moungo Mbile Sandji2 Lolo Fako LITTORAL Lekie Kadei Douala Mefou-et-Afamba Mbombete Wouri Yola Refugee Camp Sanaga-Maritime Yaounde Mfoundi Nyong-et-Mfoumou EST Refugee Center Nyong-et-Kelle Mefou-et-Akono Ngari-singo Refugee Location Mboy Haut-Nyong Refugee Urban Nyong-et-So Location UNHCR Country Ocean Office Mvila SUD Dja-Et-Lobo Boumba-Et-Ngoko Bela UNHCR Sub-Office Libongo UNHCR Field Office Vallee-du-Ntem UNHCR Field Unit Region Boundary Departement boundary REPUBLIC OF Major roads EQUATORIAL GABON Minor roads THE CONGO GUINEA 50km The boundaries and names shown and the designations used on this map do not imply official endorsement or acceptance by the United Nations. -

Morphological Characterization of Four Leguminous Crops Cultivated in Two Agro Ecological Zone: Western and Guinean Savannah Highlands of Cameroon
European Scientific Journal March 2019 edition Vol.15, No.9 ISSN: 1857 – 7881 (Print) e - ISSN 1857- 7431 Morphological Characterization of Four Leguminous Crops Cultivated in Two Agro Ecological Zone: Western and Guinean Savannah Highlands of Cameroon Momo Wobeng Nelly Blondelle, Department of Biological Sciences, Laboratory of Biodiversity and Sustainable Development, University of Ngaoundere, Cameroon Megueni Clautilde, Head of Department of Biological Sciences, University of Ngaoundere Cameroon Mandou Mouncharou Marie Solange, Department of Crop Science, FASA, University of Dschang, Cameroon Madou Chantal, Department of Biological Sciences, University of Ngaoundere, Cameroon. Institute of Agricultural Research for Development (IRAD), Garoua, Cameroon Mapongmetsem Pierre Marie, Head of Laboratory of Biodiversity and Sustainable Development, University of Ngaoundere, Cameroon Doi: 10.19044/esj.2019.v15n9p389 URL:http://dx.doi.org/10.19044/esj.2019.v15n9p389 Abstract In 2014, quarter (¼) of Africa populations were estimated to be suffering from hunger. In Cameroon, the demand for food is increasing while productivity was decreasing subsequently the food insufficiency. To solve this problem, leguminous crops were valorized in Cameroon. They represent a major source of protein especially among the poorest population, and are rich in essential amino acids such as lysine, supplementing thus the nutritional value of cereal and tuber diets. Nevertheless, their production faced the problem of soil degradation and loss of soil fertility. The main objective of this work was to characterize leguminous crops cultivated in two agro ecological zones of Cameroon. Investigations based on farmer’s knowledge on leguminous crops cultivation were done in Western highlands and High Guinean savannah zones of Cameroon. During this survey, leguminous crops were collected for qualitative and quantitative analysis. -

Prevalence and Husbandry-Related Risk Factors of Myiasis in Domestic Cavies in the Western Highlands of Cameroon
Epidemiol. Infect. (2017), 145, 339–346. © Cambridge University Press 2016 doi:10.1017/S0950268816002466 Prevalence and husbandry-related risk factors of myiasis in domestic cavies in the western highlands of Cameroon M. K. KOUAM1,2*, F. MEUTCHIEYE1,E.MIEGOUE1,T.T.NGUAFACK1, 1 1 J. TCHOUMBOUE AND A TEGUIA 1 Department of Animal Production, Faculty of Agronomy and Agricultural Sciences, Dschang, Cameroon 2 Center for Research on Filariasis and other Tropical Diseases (CRFilMT), Yaoundé, Cameroon Received 27 March 2016; Final revision 13 September 2016; Accepted 28 September 2016; first published online 26 October 2016 SUMMARY The presence of parasites on the farm can be a cause of losses in animal production, and often a threat to public health. A cross-sectional study was carried out in rural areas of the western highlands of Cameroon to determine the prevalence and husbandry-related risk factors associated with Cordylobia anthropophaga infestations in domestic cavies. The overall prevalence of myiasis in animals was 2·80% [95% confidence interval (CI) 1·50–5·10]; myiasis was found in 2% and 4·30% animals in Menoua and Bamboutos divisions, respectively. Eleven farms (8·95%) in total were infested with C. anthropophaga, with 6·41% and 13·34% of farms in the Menoua and Bamboutos divisions, respectively. The relative risk of infestation within each factor showed that the risk of myiasis in animals kept in kitchen compartments without litter was 6·16 times higher (95% CI 1·71–22·29, P = 0·04) than in animals kept in kitchens and house floors. Despite the low prevalence, the burden of cordylobiasis needs to be assessed.