January, August & Final Reports
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Draft Pest Categorisation of Organisms Associated with Washed Ware Potatoes (Solanum Tuberosum) Imported from Other Australian States and Territories
Nucleorhabdovirus Draft pest categorisation of organisms associated with washed ware potatoes (Solanum tuberosum) imported from other Australian states and territories This page is intentionally left blank Contributing authors Bennington JMA Research Officer – Biosecurity and Regulation, Plant Biosecurity Hammond NE Research Officer – Biosecurity and Regulation, Plant Biosecurity Poole MC Research Officer – Biosecurity and Regulation, Plant Biosecurity Shan F Research Officer – Biosecurity and Regulation, Plant Biosecurity Wood CE Technical Officer – Biosecurity and Regulation, Plant Biosecurity Department of Agriculture and Food, Western Australia, December 2016 Document citation DAFWA 2016, Draft pest categorisation of organisms associated with washed ware potatoes (Solanum tuberosum) imported from other Australian states and territories. Department of Agriculture and Food, Western Australia, South Perth. Copyright© Western Australian Agriculture Authority, 2016 Western Australian Government materials, including website pages, documents and online graphics, audio and video are protected by copyright law. Copyright of materials created by or for the Department of Agriculture and Food resides with the Western Australian Agriculture Authority established under the Biosecurity and Agriculture Management Act 2007. Apart from any fair dealing for the purposes of private study, research, criticism or review, as permitted under the provisions of the Copyright Act 1968, no part may be reproduced or reused for any commercial purposes whatsoever -
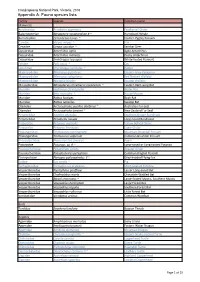
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus -

Assessing the Invertebrate Fauna Trajectories in Remediation Sites of Winstone Aggregates Hunua Quarry in Auckland
ISSN: 1179-7738 ISBN: 978-0-86476-417-1 Lincoln University Wildlife Management Report No. 59 Assessing the invertebrate fauna trajectories in remediation sites of Winstone Aggregates Hunua quarry in Auckland by Kate Curtis1, Mike Bowie1, Keith Barber2, Stephane Boyer3 , John Marris4 & Brian Patrick5 1Department of Ecology, Lincoln University, PO Box 85084, Lincoln 7647 2Winstone Aggregates, Hunua Gorge Road, Red Hill 2110, Auckland 3Department of Nature Sciences, Unitec Institute of Technology, PO Box 92025, Auckland 1142. 4Bio-Protection Research Centre, Lincoln University, PO Box 85084, Lincoln 7647. 5Consultant Ecologist, Wildlands, PO Box 33499, Christchurch. Prepared for: Winstone Aggregates April 2016 Table of Contents Abstract……………………………………………………………………………………....................... 2 Introduction…………………………………………………………………………………………………… 2 Methodology…………………………………………………………………………………………………. 4 Results…………………………………………………………………………………………………………… 8 Discussion……………………………………………………………………………………………………. 31 Conclusion…………………………………………………………………………………………………… 37 Recommendations………………………………………………………………………………………. 38 Acknowlegdements……………………………………………………………………………………… 38 References…………………………………………………………………………………………………… 39 Appendix……………………………………………………………………………………………………… 43 1 Abstract This study monitored the invertebrates in restoration plantings in the Winstone Aggregates Hunua Quarry. This was to assess the re-establishment of invertebrates in the restoration planting sites and compare them with unplanted control and mature sites. This study follows on from -

Getting the Best from Old Man Saltbush
AGFACTS Getting the best AGFACTS from old man AGFACTS saltbush Agfact P2.5.43, First edition Brett M Honeysett, Research Agronomist, Peter L Milthorpe, Senior Research Agronomist, Margaret J Wynne, Clerical Officer, Agricultural Research & Advisory Station, Condobolin INTRODUCTION Conflicting perceptions about the value of OMSB to the pastoral and farming industries have developed in Awareness is growing of the need for perennials to be recent times and these need resolution. Disappointing re-introduced into the landscape, to stem, or reverse, experiences with OMSB usually stem from unrealistic the rate of land degradation. However, reintroduction expectations of the potential of OMSB or the level of perennials into agriculture will not occur unless they of management that prevailed. have the potential to provide positive financial returns or demonstrate significant positive environmental This Agfact outlines the potential of OMSB for each benefits. Forage shrubs are a group of plants that of the different functions that it can fulfil in its offer this potential, particularly old man saltbush capacity as an environmental or productive plant. It is (OMSB). important that the reader is focussed on their reason (or reasons) for wanting to establish plantings well before commencing operations, particularly if cost recovery is important. ORDER NO. P2.5.43 AGDEX 136/10 requires an addition of a high-energy feed source to the diet if maximum production is to be achieved, especially if the leaf has a high salt content. This can be in the form of good quality grass pasture or grain. ABOUT OMSB Suitable growing areas OMSB appears highly suited to a broad region through central NSW (Figure 2). -

Charles Darwin, Kadji Kadji, Karara, Lochada Reserves WA
BUSH BLITZ SPECIES DISCOVERY PROGRAM Charles Darwin Reserve WA 3–9 May · 14–25 September · 7–18 December 2009 Kadji Kadji, Karara, Lochada Reserves WA 14–25 September · 7–18 December 2009 What is Contents Bush Blitz? Bush Blitz is a four-year, What is Bush Blitz 2 multi-million dollar Summary 3 partnership between the Abbreviations 3 Australian Government, Introduction 4 BHP Billiton, and Earthwatch Reserves Overview 5 Australia to document plants Methods 8 and animals in selected properties across Australia’s Results 10 National Reserve System. Discussion 12 Appendix A: Species Lists 15 Fauna 16 This innovative partnership Vertebrates 16 harnesses the expertise of many Invertebrates 25 of Australia’s top scientists from Flora 48 museums, herbaria, universities, Appendix B: Rare and Threatened Species 79 and other institutions and Fauna 80 organisations across the country. Flora 81 Appendix C: Exotic and Pest Species 83 Fauna 84 Flora 85 2 Bush Blitz survey report Summary Bush Blitz fieldwork was conducted at four National Reserve System properties in the Western Australian Avon Wheatbelt and Yalgoo Bioregions during 2009. This included a pilot study Abbreviations at Charles Darwin Reserve and a longer study of Charles Darwin, Kadji Kadji, Lochada and Karara reserves. Results include 651 species added to those known across the reserves and the discovery of 35 putative species new to science. The majority of ANHAT these new species occur within the heteroptera (plant bugs) and Australian Natural Heritage Assessment lepidoptera (butterflies and moths) taxonomic groups. Tool Malleefowl (Leipoa ocellata), listed as vulnerable under the EPBC Act federal Environmental Protection and Biodiversity Conservation Environment Protection and Biodiversity Act 1999 (EPBC Act), were observed on Charles Darwin Reserve. -

WO 2017/214476 Al O
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date W O 2017/214476 A l 14 December 2017 (14.12.2017) W ! P O PCT (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C12N 15/87 (2006.01) C12R 1/01 (2006.01) kind of national protection available): AE, AG, AL, AM, A01K 67/033 {2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (21) International Application Number: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, PCT/US20 17/036693 HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, (22) International Filing Date: KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, 09 June 2017 (09.06.2017) MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (25) Filing Language: English SC, SD, SE, SG, SK, SL, SM, ST, SV, SY,TH, TJ, TM, TN, (26) Publication Langi English TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 62/347,818 09 June 2016 (09.06.2016) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (71) Applicants: VANDERBILT UNIVERSITY [US/US]; UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 2201 West End Avenue, 305 Kirkland Hall, Nashville, Ten TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, nessee 37235 (US). -

Indigenous Insect Fauna and Vegetation of Rakaia Island
Indigenous insect fauna and vegetation of Rakaia Island Report No. R14/60 ISBN 978-1-927299-84-2 (print) 978-1-927299-86-6 (web) Brian Patrick Philip Grove June 2014 Report No. R14/60 ISBN 978-1-927299-84-2 (print) 978-1-927299-86-6 (web) PO Box 345 Christchurch 8140 Phone (03) 365 3828 Fax (03) 365 3194 75 Church Street PO Box 550 Timaru 7940 Phone (03) 687 7800 Fax (03) 687 7808 Website: www.ecan.govt.nz Customer Services Phone 0800 324 636 Indigenous insect fauna and vegetation of Rakaia Island Executive summary The northern end of Rakaia Island, a large in-river island of the Rakaia River, still supports relatively intact and extensive examples of formerly widespread Canterbury Plains floodplain and riverbed habitats. It is managed as a river protection reserve and conservation area by Canterbury Regional Council, having been retired from grazing since 1985. This report describes the insect fauna associated with indigenous and semi-indigenous forest, shrubland-grassland and riverbed vegetation of north Rakaia Island. A total of 119 insect species of which 112 (94%) are indigenous were recorded from the area during survey and sampling in 2012-13. North Rakaia Island is of very high ecological significance for its remnant indigenous vegetation and flora (including four nationally threatened plant species), its insect communities, and insect-plant relationships. This survey, which focused on Lepidoptera, found many of the common and characteristic moths and butterflies that would have been abundant across the Canterbury Plains before European settlement. Three rare/threatened species and several new species of indigenous moth were also found. -

Diversität Von Nachtfaltergemeinschaften Entlang Eines Höhengradienten in Südecuador (Lepidoptera: Pyraloidea, Arctiidae)
Diversität von Nachtfaltergemeinschaften entlang eines Höhengradienten in Südecuador (Lepidoptera: Pyraloidea, Arctiidae) Dissertation zur Erlangung des Doktorgrades an der Fakultät Biologie/Chemie/Geowissenschaften der Universität Bayreuth vorgelegt von Dirk Süßenbach aus Bayreuth Bayreuth, Januar 2003 Die vorliegende Arbeit wurde am Lehrstuhl Tierökologie I der Universität Bayreuth in der Arbeitsgruppe von Prof. Dr. Konrad Fiedler erstellt und von der Deutschen Forschungsgemeinschaft gefördert (Projekt Fi 547/5-1, 5-3, FOR 402/1-1 Tp 15). Vollständiger Abdruck der von der Fakultät Biologie/Chemie/Geowissenschaften der Universität Bayreuth genehmigten Dissertation zur Erlangung des Grades eines Doktors der Naturwissenschaften (Dr. rer. nat.). Tag der Einreichung: 08.01.2003 Tag des wissenschaftlichen Kolloquiums: 09.04.2003 1. Gutachter: Prof. Dr. K. Fiedler 2. Gutachter: PD Dr. B. Stadler Prüfungsausschuss: Prof. Dr. G. Rambold (Vorsitzender) Prof. Dr. K. Dettner Prof. Dr. Chr. Engels INHALTSVERZEICHNIS 1. Einleitung ...................................................................................................................... 1 2. Untersuchungsgebiet ................................................................................................... 13 3. Methodik...................................................................................................................... 20 3.1 Lichtfang.............................................................................................................. 20 3.2 Probennahme, Präparation -

Olabisi Onabanjo University, Ago Iwoye, P.M.B
. ISSN: 1817-3098 © International Digital Organization for Scientific Information Online Open Access Volume 1 Number(1) : 40-53, Jan-Jun, 2006 Insects Associated with Some Medicinal Plants in South-Western Nigeria A.D. Banjo, O.A. Lawal and S.A. Aina Department of Plant Science and Applied Zoology, Olabisi Onabanjo University, Ago Iwoye, P.M.B. 2002, Ogun State, Nigeria Abstract: Insects associated with Vernonia amygdalina, Bryophyllum pinnatum, Boerheavia diffusa, Spondias mombin, Gliricidia sepium and Momordica charantia were surveyed. Insects found on the plants parts include Diptera, Hymenoptera, Coleoptera, Orthoptera, Lepidoptera, Dictyoptera, Homoptera and Isoptera. The species richness, diversity index and the similarly index between paired plant species were also calculated. The results of similarity and diversity indexes were influenced by the weather, which fluctuated during the period of study. The Hymenoptera was the most abundant order on Vernonia amygdalina, Boerheavia diffusa, Spondias mombin, Gliricidia sepium and Momordica charantia accounting for 44.6, 64.5, 45.2, 56.4 and 42.02%, respectively. Diptera occur the most on Brophyllum pinnatum with 40.4% share of the total population. Key words: Medicinal plants insects species richness South-western Nigeria INTRODUCTION local medicine as antiparasitic, antipyretic, laxative and tonic [4]. Medicinal plants are of great importance to man as The leaves are rubbed on the body for itching conditions, alternative for medicinal purposes, also used as food sources and parasitic skin diseases and ringworm etc. Women drink a can be put into use as pesticides in form of organic derivatives decoction of the leaves to affect the milk and act as a vicarious or synthetics. -

I SPY Section 4
This document is part of a larger publication “I Spy – Insects of Southern Australian Broadacre Farming Systems Identification Manual and Educational Resource” (ISBN 978-0-6482692-1-2) produced under the National Invertebrate Pest Initiative (NIPI) and is subject to the disclaimers and copyright of the full version from which it was extracted. The remaining sections and the full version of this publication, as well as updates and other legal information, can be found at https://grdc.com.au/ by searching for “I SPY”. 2018 © The development of this edition of I SPY has been possible due to the financial support from: Insects of Southern Australian Broadacre Farming Systems Identification Manual and Education Resource Education Resource Manual and Identification Systems Farming Insects Broadacre of Southern Australian SECTION 4 Common Pest, Beneficial and Exotic Species Moths & Butterflies . 2 Armyworms . 4 Legend Cutworms . 7 Budworms . 11 Crop type Diamondback moth . 13 Lucerne seed web moth . 15 Cereals Beetles . 17 Cockchafers . 19 Canola (& Brassicas) True wireworms . 24 False wireworms . 26 Pulses & legumes Weevils . 28 Ladybird beetles . 31 Pasture Carabid beetles (or Ground beetles) . 33 Bugs . 35 Monitoring type Figure 4.1 An example of an aphid lifecycle (Aphis sp.) . 37 Figure 4.2 Persistent versus non-persistent transmission of viruses . 38 Observation Table 4.1 Some aphids known to transmit viruses in pulse crops . 38 Cereal aphids - Corn aphid, Oat aphid & Russian wheat aphid . 39 Sweep net Canola aphids - Cabbage aphid, Turnip aphid & Green peach aphid . 42 Pulse aphids - Blue green aphid, Pea aphid, Cow-pea aphid & Yellow sticky trap Green peach aphid . -

Mt Canobolas SCA AOBV Nomination – Medd & Bower - 2
Nomination of Mount Canobolas State Conservation Area as an Area of Outstanding Biodiversity Value 1Richard W. Medd PhD and 2Colin C. Bower PhD 1593 Cargo Road Orange, NSW 2800 [email protected] 2 PO Box 300, Orange, NSW 2800 Original Submitted May 2018 Revised with minor corrections July 2018 Citation: Medd RW and Bower CC (2018). Nomination of Mount Canobolas State Conservation Area as an Area of Outstanding Biodiversity Value. Submission to Office of Environment and Heritage, Unpublished 63pp Mt Canobolas SCA AOBV Nomination – Medd & Bower - 2 - Table of Contents Page Executive Summary ...................................................................................................... 3 1. Introduction ......................................................................................................... 7 2. Background ......................................................................................................... 7 3. Bioheritage .......................................................................................................... 8 3.1 Bryophytes 3.2 Vascular plants 3.3 Fungi 3.4 Vertebrates 3.5 Invertebrates 4. Threatened Ecological Communities .................................................................. 11 4.1 Xanthoparmelia Lichen Community 4.2 Tableland Basalt Forest Community 4.3 Tablelands Snow Gum Woodland Community 5. Threatened Species ............................................................................................ 12 5.1 Plants 5.2 Mammals 5.3 Birds 6. Endemic Species ............................................................................................... -

Gwambygine Pool Conservation Reserve
Gwambygine Pool Conservation Reserve Fauna (macro-invertebrate and herpetofauna) Inventory Survey - 2009/10 ©David Knowles Spineless Wonders Fauna Surveys Prepared and published for the River Conservation Society York Western Australia with financial assistance from Wheatbelt Natural Resource Management. Cover Photograph: Common male Gwambygine Pool Damselfies (above) Blue Ringtail Lestidamselfly Austrolestes annulosus (centre) Redfront Coenadamselfly Xanthagrion erythroneurum (below) Aurora Bluetail Coenadamslefly Ischnura heterosticta Report David Knowles Design Fleur Knowles Spineless Wonders 08 9247 5772 [email protected] www.spinelesswonders.com.au CONTENTS Page Forward 4 Introduction 5 Maps and Pie Graph 9 Scope 12 Specimen preservation and collection 13 Voucher Specimen Lodgement 13 Interpretation aids 13 DEC Permit 13 Sampling Methods 14 Definitions 16 Site Subregion and Description 17 Plants of the Immediate Riparian Zone 18 Results - Ordinal Summaries and Tables 19 Conclusions 44 Recommendations 45 Acknowledgements 46 Web Resources 46 References 47 Interpreting The Tables – DefinitionsSymbols 48 Text And Abbreviations 48 MAIN TABLE APPENDIX 1 - 6 49 Table Appendix 7 - Table Of Commom Naming Codes 102 - 124 COLOUR SUPPLEMENT Spring (GREEN) 1 Summer (RED) 30 Autumn (GOLD) 58 Gwambygine Pool Conservation Reserve Fauna (macro-invertebrate and herpetofauna) Inventory Survey Forward It gives me a great deal of pleasure to present this Invertebrate Survey of Gwambygine Pool Conservation Reserve by David Knowles. The River Conservation Society commissioned David to carry out three surveys of the Reserve in September, October and November 2009 and in May 2010. The survey has revealed a rich diversity and abundance of the terrestrial invertebrate population in close proximity to Gwambygine Pool – a much greater abundance than had been expected.