OR4F16 Antibody Cat
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tumor Elastography and Its Association with Cell-Free Tumor DNA in the Plasma of Breast Tumor Patients: a Pilot Study
3534 Original Article Tumor elastography and its association with cell-free tumor DNA in the plasma of breast tumor patients: a pilot study Yi Hao1#, Wei Yang2#, Wenyi Zheng2,3#, Xiaona Chen3,4, Hui Wang1,5, Liang Zhao1,5, Jinfeng Xu6,7, Xia Guo4 1Department of Ultrasound, South China Hospital of Shenzhen University, Shenzhen, China; 2Department of Ultrasound, Shenzhen Hospital, Southern Medical University, Shenzhen, China; 3The Third School of Clinical Medicine, Southern Medical University, Guangzhou, China; 4Shenzhen Key Laboratory of Viral Oncology, Center for Clinical Research and Innovation (CCRI), Shenzhen Hospital, Southern Medical University, Shenzhen, China; 5Department of Ultrasound, Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, China; 6Department of Ultrasound, Shenzhen People’s Hospital (The Second Clinical Medical College, Jinan University, Shenzhen, China; 7The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen, China #These authors contributed equally to this work. Correspondence to: Xia Guo. Shenzhen Key Laboratory of Viral Oncology, Center for Clinical Research and Innovation (CCRI), Shenzhen Hospital, Southern Medical University, Shenzhen 518000, China. Email: [email protected]; Jinfeng Xu. Department of Ultrasound, Shenzhen People’s Hospital (The Second Clinical Medical College, Jinan University, Shenzhen 518020, China; The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen 518020, China. Email: [email protected]. Background: Breast tumor stiffness, which can be objectively and noninvasively evaluated by ultrasound elastography (UE), has been useful for the differentiation of benign and malignant breast lesions and the prediction of clinical outcomes. Liquid biopsy analyses, including cell-free tumor DNA (ctDNA), exhibit great potential for personalized treatment. This study aimed to investigate the correlations between the UE and ctDNA for early breast cancer diagnosis. -
Quantitative-PCR Validation of 154 Genomic Segments Called As Cnvs in Five Replicat
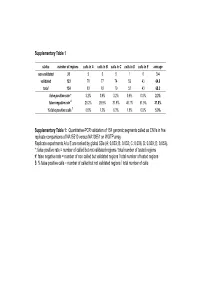
Supplementary Table 1 status number of regions calls in A calls in B calls in C calls in D calls in E average non validated 31 5 6 5 1 0 3.4 validated 123 78 77 74 52 43 64.8 total 154 83 83 79 53 43 68.2 false positive rate * 3.2% 3.9% 3.2% 0.6% 0.0% 2.2% false negative rate # 29.2% 29.9% 31.8% 46.1% 51.9% 37.8% % false positive calls $ 6.0% 7.2% 6.3% 1.9% 0.0% 5.0% Supplementary Table 1: Quantitative-PCR validation of 154 genomic segments called as CNVs in five replicate comparisons of NA15510 versus NA10851 on WGTP array Replicate experiments A to E are ranked by global SDe (A: 0.033; B: 0.033; C: 0.036; D: 0.039; E: 0.053). *: false positive rate = number of called but not validated regions / total number of tested regions #: false negative rate = number of non called but validated regions / total number of tested regions $: % false positive calls = number of called but not validated regions / total number of calls False positive estimates for 500K EA CNV calls Total Rep1 Rep2 Rep3 Avg (unique) Validated 33 28 32 31 38 Not validated 2 2 2 2 5 Total 35 30 34 33 43 % False positive 5.71% 6.67% 5.88% 6.09% - % False negative 13.16% 26.32% 15.79% 18.42% - Supplementary Table 2A : Quantitative PCR validation of 43 unique CNV regions called as CNVs in three replicate comparisons of NA15510 versus NA10851 using the 500K EA array. -

Output Results of CLIME (Clustering by Inferred Models of Evolution)
Output results of CLIME (CLustering by Inferred Models of Evolution) Dataset: Num of genes in input gene set: 4 Total number of genes: 20834 Prediction LLR threshold: 0 The CLIME PDF output two sections: 1) Overview of Evolutionarily Conserved Modules (ECMs) Top panel shows the predefined species tree. Bottom panel shows the partition of input genes into Evolutionary Conserved Modules (ECMs), ordered by ECM strength (shown at right), and separated by horizontal lines. Each row show one gene, where the phylogenetic profile indicates presence (blue) or absence (gray) of homologs in each species (column). Gene symbols are shown at left. Gray color indicates that the gene is a paralog to a higher scoring gene within the same ECM (based on BLASTP E < 1e-3). 2) Details of each ECM and its expansion ECM+ Top panel shows the inferred evolutionary history on the predefined species tree. Branch color shows the gain event (blue) and loss events (red color, with brighter color indicating higher confidence in loss). Branches before the gain or after a loss are shown in gray. Bottom panel shows the input genes that are within the ECM (blue/white rows) as well as all genes in the expanded ECM+ (green/gray rows). The ECM+ includes genes likely to have arisen under the inferred model of evolution relative to a background model, and scored using a log likelihood ratio (LLR). PG indicates "paralog group" and are labeled alphabetically (i.e., A, B). The first gene within each paralog group is shown in black color. All other genes sharing sequence similarity (BLAST E < 1e-3) are assigned to the same PG label and displayed in gray. -

Epigenetic Trajectories to Childhood Asthma
Epigenetic Trajectories to Childhood Asthma Item Type text; Electronic Dissertation Authors DeVries, Avery Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction, presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 04/10/2021 09:24:24 Link to Item http://hdl.handle.net/10150/630233 1 EPIGENETIC TRAJECTORIES TO CHILDHOOD ASTHMA by Avery DeVries __________________________ Copyright © Avery DeVries 2018 A Dissertation Submitted to the Faculty of the DEPARTMENT OF CELLULAR AND MOLECULAR MEDICINE In Partial Fulfillment of the Requirements For the Degree of DOCTOR OF PHILOSOPHY In the Graduate College THE UNIVERSITY OF ARIZONA 2018 2 3 STATEMENT BY AUTHOR This dissertation has been submitted in partial fulfillment of the requirements for an advanced degree at the University of Arizona and is deposited in the University Library to be made available to borrowers under rules of the Library. Brief quotations from this dissertation are allowable without special permission, provided that an accurate acknowledgement of the source is made. Requests for permission for extended quotation from or reproduction of this manuscript in whole or in part may be granted by the head of the major department or the Dean of the Graduate College when in his or her judgment the proposed use of the material is in the interests of scholarship. In all other instances, however, permission must be obtained from the author. SIGNED: Avery DeVries 4 ACKNOWLEDGEMENTS I would like to thank my dissertation mentor, Donata Vercelli, for endless support throughout this journey and for always encouraging and challenging me to be a better scientist. -

Us 2018 / 0305689 A1
US 20180305689A1 ( 19 ) United States (12 ) Patent Application Publication ( 10) Pub . No. : US 2018 /0305689 A1 Sætrom et al. ( 43 ) Pub . Date: Oct. 25 , 2018 ( 54 ) SARNA COMPOSITIONS AND METHODS OF plication No . 62 /150 , 895 , filed on Apr. 22 , 2015 , USE provisional application No . 62/ 150 ,904 , filed on Apr. 22 , 2015 , provisional application No. 62 / 150 , 908 , (71 ) Applicant: MINA THERAPEUTICS LIMITED , filed on Apr. 22 , 2015 , provisional application No. LONDON (GB ) 62 / 150 , 900 , filed on Apr. 22 , 2015 . (72 ) Inventors : Pål Sætrom , Trondheim (NO ) ; Endre Publication Classification Bakken Stovner , Trondheim (NO ) (51 ) Int . CI. C12N 15 / 113 (2006 .01 ) (21 ) Appl. No. : 15 /568 , 046 (52 ) U . S . CI. (22 ) PCT Filed : Apr. 21 , 2016 CPC .. .. .. C12N 15 / 113 ( 2013 .01 ) ; C12N 2310 / 34 ( 2013. 01 ) ; C12N 2310 /14 (2013 . 01 ) ; C12N ( 86 ) PCT No .: PCT/ GB2016 /051116 2310 / 11 (2013 .01 ) $ 371 ( c ) ( 1 ) , ( 2 ) Date : Oct . 20 , 2017 (57 ) ABSTRACT The invention relates to oligonucleotides , e . g . , saRNAS Related U . S . Application Data useful in upregulating the expression of a target gene and (60 ) Provisional application No . 62 / 150 ,892 , filed on Apr. therapeutic compositions comprising such oligonucleotides . 22 , 2015 , provisional application No . 62 / 150 ,893 , Methods of using the oligonucleotides and the therapeutic filed on Apr. 22 , 2015 , provisional application No . compositions are also provided . 62 / 150 ,897 , filed on Apr. 22 , 2015 , provisional ap Specification includes a Sequence Listing . SARNA sense strand (Fessenger 3 ' SARNA antisense strand (Guide ) Mathew, Si Target antisense RNA transcript, e . g . NAT Target Coding strand Gene Transcription start site ( T55 ) TY{ { ? ? Targeted Target transcript , e . -
Explorations in Olfactory Receptor Structure and Function by Jianghai
Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 ABSTRACT Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 Copyright by Jianghai Ho 2014 Abstract Olfaction is one of the most primitive of our senses, and the olfactory receptors that mediate this very important chemical sense comprise the largest family of genes in the mammalian genome. It is therefore surprising that we understand so little of how olfactory receptors work. In particular we have a poor idea of what chemicals are detected by most of the olfactory receptors in the genome, and for those receptors which we have paired with ligands, we know relatively little about how the structure of these ligands can either activate or inhibit the activation of these receptors. Furthermore the large repertoire of olfactory receptors, which belong to the G protein coupled receptor (GPCR) superfamily, can serve as a model to contribute to our broader understanding of GPCR-ligand binding, especially since GPCRs are important pharmaceutical targets. -

Novel Cell Lines and Methods
(19) TZZ¥ZZ_¥_T (11) EP 3 009 513 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 20.04.2016 Bulletin 2016/16 C12N 15/85 (2006.01) C12N 15/67 (2006.01) C07K 14/435 (2006.01) (21) Application number: 15180871.4 (22) Date of filing: 02.02.2009 (84) Designated Contracting States: (72) Inventor: SHEKDAR, Kambiz AT BE BG CH CY CZ DE DK EE ES FI FR GB GR New York, NY 10010 (US) HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL PT RO SE SI SK TR (74) Representative: Vossius & Partner Patentanwälte Rechtsanwälte mbB (30) Priority: 01.02.2008 US 63219 P Siebertstrasse 3 81675 München (DE) (62) Document number(s) of the earlier application(s) in accordance with Art. 76 EPC: Remarks: 09709529.3 / 2 245 171 •Claims filed after the date of filing of the application (Rule 68(4) EPC). (71) Applicant: Chromocell Corporation •This application was filed on 13-08-2015 as a North Brunswick, NJ 08902 (US) divisional application to the application mentioned under INID code 62. (54) NOVEL CELL LINES AND METHODS (57) The invention relates to novel cells and cell lines, and methods for making and using them. EP 3 009 513 A1 Printed by Jouve, 75001 PARIS (FR) EP 3 009 513 A1 Description Field of the Invention 5 [0001] The invention relates to novel cells and cell lines, and methods for making and using them. Background of the Invention [0002] Currently, the industry average failure rate for drug discovery programs in pharmaceutical companies is reported 10 to be approximately 98%. -
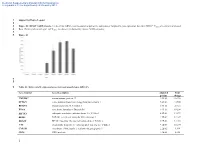
Supporting Figure Legend Figure S1
Electronic Supplementary Material (ESI) for Metallomics This journal is © The Royal Society of Chemistry 2013 1 Supporting Figure Legend 2 3 Figure S1: RPS4Y1 mRNA levels. Levels of this mRNA were measured as part of the validation of Affymetrix gene expression data from DEG.C. Cclones are controls (untreated 4 Beas-2B that grew in soft agar) and Vclones are clones transformed by chronic NaVO3 exposure. 5 6 Figure S1 7 8 9 Table S1: Differentially expressed genes in transformed clones (DEG.C). Gene Symbol Gene Description Adjusted Fold p-value change TMEM47 transmembrane protein 47 1.9E-02 36.196 SPTLC3 serine palmitoyltransferase, long chain base subunit 3 7.8E-03 33.565 RPS4Y1 ribosomal protein S4, Y-linked 1 6.9E-03 28.724 EYA4 eyes absent homolog 4 (Drosophila) 3.9E-03 18.298 EIF1AY eukaryotic translation initiation factor 1A, Y-linked 8.6E-03 13.872 EDIL3 EGF-like repeats and discoidin I-like domains 3 1.3E-02 12.147 DDX3Y DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked 3.9E-03 12.110 UTY ubiquitously transcribed tetratricopeptide repeat gene, Y-linked 9.5E-03 10.134 CYP1B1 cytochrome P450, family 1, subfamily B, polypeptide 1 2.2E-02 9.194 CD24 CD24 molecule 1.4E-02 8.328 1 Electronic Supplementary Material (ESI) for Metallomics This journal is © The Royal Society of Chemistry 2013 GABRA3 gamma-aminobutyric acid (GABA) A receptor, alpha 3 1.7E-02 7.643 ZNF730 zinc finger protein 730 2.0E-02 7.283 RARB|LOC100130354 retinoic acid receptor, beta | hypothetical protein LOC100130354 3.3E-02 7.168 DNER delta/notch-like EGF repeat containing -

Supplementary Figure 1 1 2 Promoter Analysis for the Putative
1 Supplementary Figure 1 2 3 Promoter analysis for the putative BRCA1 target genes. (A) A graph for luciferase reporter assay of 4 the CKS1B and MEIS2 promoter plasmids, in combination with wild type BRCA1 (wtBRCA1) or non- 5 functional mutant (C61G). (B) ChIP results for CKS1B and MEIS2 promoter with two different BRCA1 6 antibodies and HDAC2. IgG and Input was used as a negative or positive control, respectively. 7 8 9 Supplementary Table 1. The BRCA1 ChIP signal from ENCODE database and BRCA1 dependent expression change from two GEO datasets GSE22259 GSE30822 Gene Name ENCODE data (BRCA1 CHIP seq) Expression Expression data data ANPEP unnamed (-79179) #N/A 0.140046483 CENPE unnamed (-76) 0.463340941 0.155474015 ID1 unnamed (-32379), unnamed (+7707) 1.020982905 0.214958903 PNN unnamed (+161) 0.772059373 0.215856379 SUSD2 unnamed (-24295) #N/A 0.230809307 LEMD1 unnamed (-63610) #N/A 0.254516474 DLGAP5 unnamed (-79580) 0.914187251 0.281355563 DARS2 unnamed (-11) 0.9045215 0.289088038 SMC3 unnamed (-361) 1.021103204 0.309860113 TNNI3 unnamed (-3119) #N/A 0.318065991 FUT8 unnamed (+1870) 0.992421144 0.322867733 MOB1B unnamed (-154) #N/A 0.328830217 HMGCS1 unnamed (-283) 1.677479154 0.338263784 CTH unnamed (-163) 1.30602241 0.341207388 SEPT10 unnamed (+15) 1.04077455 0.345515568 SLC45A3 unnamed (-68696), unnamed (+49304) #N/A 0.346195509 TAF1B unnamed (-73) 0.797296768 0.348470135 OXCT1 unnamed (-33516) 0.996978985 0.353844424 KIF15 unnamed (+76) 0.678047851 0.356200772 FBP1 unnamed (-87049) #N/A 0.36391027 C4orf21 unnamed (-234) #N/A 0.367754534 -

Distinct Patterns of Epigenetic Marks and Transcription Factor Binding Sites Across Promoters of Sense-Intronic Long Noncoding Rnas
Supplementary data: Distinct patterns of epigenetic marks and transcription factor binding sites across promoters of sense-intronic long noncoding RNAs Sourav Ghosh, Satish Sati, Shantanu Sengupta and Vinod Scaria J. Genet. 94, 17–25 Gencode V9 lncRNA gene : 11004 Known lncRNA : 1175 Novel lncRNA : 5898 Putative lncRNA : 3931 Min Max Figure 1. Different histone modifications and TFBS site distribution patterns across the TSS of known, novel and putative lncRNA genes. Histone modification peak summit count was calculated in the 100 bp sliding window within 5 kb upstream and downstream regions from TSS in H1 cell line. Peak summit count was further normalized by dividing each bin with the total number of genes in each category. Journal of Genetics Supplementary data, J. Genet. 94, 17–25 a b c Figure 2. RNA pol II and pol III occupancy around TSS, represented as heat maps. (a) RNA pol III peak summits of K562 cell types plotted across the TSS, 5 kb upstream and downstream in 100 bp nonoverlapping sliding window, for all protein coding, all long noncoding and lncRNA subclass genes. (b) RNA pol III peak summit count in 100bp non-overlapping sliding window, 5 kb upstream and downstream from the start site calculated for all protein-coding genes, lncRNA genes and lncRNA subclasses from GM12878 cell types. (c) RNA pol II peak summits calculated in 100 bp sliding window, 5 kb upstream and downstream from the TSS site was calculated for all protein coding, all lncRNA and lncRNA subclasses genes in GM12878 cell type. Count was normalized by dividing individual count with total number of genes in that category. -

Supplementary Methods
doi: 10.1038/nature06162 SUPPLEMENTARY INFORMATION Supplementary Methods Cloning of human odorant receptors 423 human odorant receptors were cloned with sequence information from The Olfactory Receptor Database (http://senselab.med.yale.edu/senselab/ORDB/default.asp). Of these, 335 were predicted to encode functional receptors, 45 were predicted to encode pseudogenes, 29 were putative variant pairs of the same genes, and 14 were duplicates. We adopted the nomenclature proposed by Doron Lancet 1. OR7D4 and the six intact odorant receptor genes in the OR7D4 gene cluster (OR1M1, OR7G2, OR7G1, OR7G3, OR7D2, and OR7E24) were used for functional analyses. SNPs in these odorant receptors were identified from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) or through genotyping. OR7D4 single nucleotide variants were generated by cloning the reference sequence from a subject or by inducing polymorphic SNPs by site-directed mutagenesis using overlap extension PCR. Single nucleotide and frameshift variants for the six intact odorant receptors in the same gene cluster as OR7D4 were generated by cloning the respective genes from the genomic DNA of each subject. The chimpanzee OR7D4 orthologue was amplified from chimpanzee genomic DNA (Coriell Cell Repositories). Odorant receptors that contain the first 20 amino acids of human rhodopsin tag 2 in pCI (Promega) were expressed in the Hana3A cell line along with a short form of mRTP1 called RTP1S, (M37 to the C-terminal end), which enhances functional expression of the odorant receptors 3. For experiments with untagged odorant receptors, OR7D4 RT and S84N variants without the Rho tag were cloned into pCI. -

Research/0018.1
http://genomebiology.com/2001/2/6/research/0018.1 Research The human olfactory receptor repertoire comment Sergey Zozulya, ernando Echeverri and Trieu Nguyen Address: Senomyx Inc., 11099 North Torrey Pines Road, La Jolla, CA 92037, USA. Correspondence: Sergey Zozulya. E-mail: [email protected] reviews Published: 1 June 2001 Received: 8 March 2001 Revised: 12 April 2001 Genome Biology 2001, 2(6):research0018.1–0018.12 Accepted: 18 April 2001 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2001/2/6/research/0018 © 2001 Zozulya et al., licensee BioMed Central Ltd (Print ISSN 1465-6906; Online ISSN 1465-6914) reports Abstract Background: The mammalian olfactory apparatus is able to recognize and distinguish thousands of structurally diverse volatile chemicals. This chemosensory function is mediated by a very large family of seven-transmembrane olfactory (odorant) receptors encoded by approximately 1,000 genes, the majority of which are believed to be pseudogenes in humans. deposited research Results: The strategy of our sequence database mining for full-length, functional candidate odorant receptor genes was based on the high overall sequence similarity and presence of a number of conserved sequence motifs in all known mammalian odorant receptors as well as the absence of introns in their coding sequences. We report here the identification and physical cloning of 347 putative human full-length odorant receptor genes. Comparative sequence analysis of the predicted gene products allowed us to identify and define a number of consensus sequence motifs and structural features of this vast family of receptors.