Supporting Online Material for Evolutionary and Biomedical Insights from the Rhesus Macaque Genome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Variation Across the Human Olfactory Receptor Repertoire Alters Odor Perception
bioRxiv preprint doi: https://doi.org/10.1101/212431; this version posted November 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genetic variation across the human olfactory receptor repertoire alters odor perception Casey Trimmer1,*, Andreas Keller2, Nicolle R. Murphy1, Lindsey L. Snyder1, Jason R. Willer3, Maira Nagai4,5, Nicholas Katsanis3, Leslie B. Vosshall2,6,7, Hiroaki Matsunami4,8, and Joel D. Mainland1,9 1Monell Chemical Senses Center, Philadelphia, Pennsylvania, USA 2Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, New York, USA 3Center for Human Disease Modeling, Duke University Medical Center, Durham, North Carolina, USA 4Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina, USA 5Department of Biochemistry, University of Sao Paulo, Sao Paulo, Brazil 6Howard Hughes Medical Institute, New York, New York, USA 7Kavli Neural Systems Institute, New York, New York, USA 8Department of Neurobiology and Duke Institute for Brain Sciences, Duke University Medical Center, Durham, North Carolina, USA 9Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA *[email protected] ABSTRACT The human olfactory receptor repertoire is characterized by an abundance of genetic variation that affects receptor response, but the perceptual effects of this variation are unclear. To address this issue, we sequenced the OR repertoire in 332 individuals and examined the relationship between genetic variation and 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. -

An Order Estimation Based Approach to Identify Response Genes
AN ORDER ESTIMATION BASED APPROACH TO IDENTIFY RESPONSE GENES FOR MICRO ARRAY TIME COURSE DATA A Thesis Presented to The Faculty of Graduate Studies of The University of Guelph by ZHIHENG LU In partial fulfilment of requirements for the degree of Doctor of Philosophy September, 2008 © Zhiheng Lu, 2008 Library and Bibliotheque et 1*1 Archives Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-47605-5 Our file Notre reference ISBN: 978-0-494-47605-5 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library permettant a la Bibliotheque et Archives and Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par Plntemet, prefer, telecommunication or on the Internet, distribuer et vendre des theses partout dans loan, distribute and sell theses le monde, a des fins commerciales ou autres, worldwide, for commercial or non sur support microforme, papier, electronique commercial purposes, in microform, et/ou autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in et des droits moraux qui protege cette these. this thesis. Neither the thesis Ni la these ni des extraits substantiels de nor substantial extracts from it celle-ci ne doivent etre imprimes ou autrement may be printed or otherwise reproduits sans son autorisation. -

Tumor Elastography and Its Association with Cell-Free Tumor DNA in the Plasma of Breast Tumor Patients: a Pilot Study
3534 Original Article Tumor elastography and its association with cell-free tumor DNA in the plasma of breast tumor patients: a pilot study Yi Hao1#, Wei Yang2#, Wenyi Zheng2,3#, Xiaona Chen3,4, Hui Wang1,5, Liang Zhao1,5, Jinfeng Xu6,7, Xia Guo4 1Department of Ultrasound, South China Hospital of Shenzhen University, Shenzhen, China; 2Department of Ultrasound, Shenzhen Hospital, Southern Medical University, Shenzhen, China; 3The Third School of Clinical Medicine, Southern Medical University, Guangzhou, China; 4Shenzhen Key Laboratory of Viral Oncology, Center for Clinical Research and Innovation (CCRI), Shenzhen Hospital, Southern Medical University, Shenzhen, China; 5Department of Ultrasound, Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, China; 6Department of Ultrasound, Shenzhen People’s Hospital (The Second Clinical Medical College, Jinan University, Shenzhen, China; 7The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen, China #These authors contributed equally to this work. Correspondence to: Xia Guo. Shenzhen Key Laboratory of Viral Oncology, Center for Clinical Research and Innovation (CCRI), Shenzhen Hospital, Southern Medical University, Shenzhen 518000, China. Email: [email protected]; Jinfeng Xu. Department of Ultrasound, Shenzhen People’s Hospital (The Second Clinical Medical College, Jinan University, Shenzhen 518020, China; The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen 518020, China. Email: [email protected]. Background: Breast tumor stiffness, which can be objectively and noninvasively evaluated by ultrasound elastography (UE), has been useful for the differentiation of benign and malignant breast lesions and the prediction of clinical outcomes. Liquid biopsy analyses, including cell-free tumor DNA (ctDNA), exhibit great potential for personalized treatment. This study aimed to investigate the correlations between the UE and ctDNA for early breast cancer diagnosis. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

OR4F16 Antibody Cat
OR4F16 Antibody Cat. No.: 57-758 OR4F16 Antibody Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This OR4F16 antibody is generated from rabbits immunized with a KLH conjugated IMMUNOGEN: synthetic peptide between 68-96 amino acids from the N-terminal region of human OR4F16. TESTED APPLICATIONS: WB APPLICATIONS: For WB starting dilution is: 1:1000 PREDICTED MOLECULAR 35 kDa WEIGHT: Properties This antibody is purified through a protein A column, followed by peptide affinity PURIFICATION: purification. CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated September 27, 2021 1 https://www.prosci-inc.com/or4f16-antibody-57-758.html PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: OR4F3 ALTERNATE NAMES: Olfactory receptor 4F3/4F16/4F29, Olfactory receptor OR1-1, OR4F3 ACCESSION NO.: Q6IEY1 PROTEIN GI NO.: 74762307 GENE ID: 26683, 729759, 81399 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Olfactory receptors interact with odorant molecules in the nose, to initiate a neuronal response that triggers the perception of a smell. The olfactory receptor proteins are members of a large family of G-protein-coupled receptors (GPCR) arising from single coding-exon genes. Olfactory receptors share a 7-transmembrane domain structure with BACKGROUND: many neurotransmitter and hormone receptors and are responsible for the recognition and G protein-mediated transduction of odorant signals. -

Valproic Acid Reversed Pathologic Endothelial Cell Gene Expression Profile Associated with Ischemia– Reperfusion Injury in a Swine Hemorrhagic Shock Model
From the Peripheral Vascular Surgery Society Valproic acid reversed pathologic endothelial cell gene expression profile associated with ischemia– reperfusion injury in a swine hemorrhagic shock model Marlin Wayne Causey, MD,a Shashikumar Salgar, PhD,b Niten Singh, MD,a Matthew Martin, MD,a and Jonathan D. Stallings, PhD,b Tacoma, Wash Background: Vascular endothelial cells serve as the first line of defense for end organs after ischemia and reperfusion injuries. The full etiology of this dysfunction is poorly understood, and valproic acid (VPA) has proven to be beneficial after traumatic injury. The purpose of this study was to determine the mechanism of action through which VPA exerts its beneficial effects. Methods: Sixteen Yorkshire swine underwent a standardized protocol for an ischemia–reperfusion injury through received (6 ؍ hemorrhage and a supraceliac cross-clamp with ensuing 6-hour resuscitation. The experimental swine (n Aortic .(5 ؍ and injury-control models (n (5 ؍ VPA at cross-clamp application and were compared with sham (n endothelium was harvested, and microarray analysis was performed along with a functional clustering analysis with gene transcript validation using relative quantitative polymerase chain reaction. Results: Clinical comparison of experimental swine matched for sex, weight, and length demonstrated that VPA significantly decreased resuscitative requirements, with improved hemodynamics and physiologic laboratory measure- ments. Six transcript profiles from the VPA treatment were compared with the 1536 gene transcripts (529 up and 1007 down) from sham and injury-control swine. Microarray analysis and a Database for Annotation, Visualization and Integrated Discovery functional pathway analysis approach identified biologic processes associated with pathologic vascular endothelial function, specifically through functional cluster pathways involving apoptosis/cell death and angiogenesis/vascular development, with five specific genes (THBS1, TNFRSF12A, ANGPTL4, RHOB, and RTN4) identified as members of both functional clusters. -

Elucidating Biological Roles of Novel Murine Genes in Hearing Impairment in Africa
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 19 September 2019 doi:10.20944/preprints201909.0222.v1 Review Elucidating Biological Roles of Novel Murine Genes in Hearing Impairment in Africa Oluwafemi Gabriel Oluwole,1* Abdoulaye Yal 1,2, Edmond Wonkam1, Noluthando Manyisa1, Jack Morrice1, Gaston K. Mazanda1 and Ambroise Wonkam1* 1Division of Human Genetics, Department of Pathology, Faculty of Health Sciences, University of Cape Town, Observatory, Cape Town, South Africa. 2Department of Neurology, Point G Teaching Hospital, University of Sciences, Techniques and Technology, Bamako, Mali. *Correspondence to: [email protected]; [email protected] Abstract: The prevalence of congenital hearing impairment (HI) is highest in Africa. Estimates evaluated genetic causes to account for 31% of HI cases in Africa, but the identification of associated causative genes mutations have been challenging. In this study, we reviewed the potential roles, in humans, of 38 novel genes identified in a murine study. We gathered information from various genomic annotation databases and performed functional enrichment analysis using online resources i.e. genemania and g.proflier. Results revealed that 27/38 genes are express mostly in the brain, suggesting additional cognitive roles. Indeed, HERC1- R3250X had been associated with intellectual disability in a Moroccan family. A homozygous 216-bp deletion in KLC2 was found in two siblings of Egyptian descent with spastic paraplegia. Up to 27/38 murine genes have link to at least a disease, and the commonest mode of inheritance is autosomal recessive (n=8). Network analysis indicates that 20 other genes have intermediate and biological links to the novel genes, suggesting their possible roles in HI. -

Identification of Candidate Biomarkers and Pathways Associated with Type 1 Diabetes Mellitus Using Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.08.447531; this version posted June 9, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of candidate biomarkers and pathways associated with type 1 diabetes mellitus using bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.06.08.447531; this version posted June 9, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Type 1 diabetes mellitus (T1DM) is a metabolic disorder for which the underlying molecular mechanisms remain largely unclear. This investigation aimed to elucidate essential candidate genes and pathways in T1DM by integrated bioinformatics analysis. In this study, differentially expressed genes (DEGs) were analyzed using DESeq2 of R package from GSE162689 of the Gene Expression Omnibus (GEO). Gene ontology (GO) enrichment analysis, REACTOME pathway enrichment analysis, and construction and analysis of protein-protein interaction (PPI) network, modules, miRNA-hub gene regulatory network and TF-hub gene regulatory network, and validation of hub genes were then performed. A total of 952 DEGs (477 up regulated and 475 down regulated genes) were identified in T1DM. GO and REACTOME enrichment result results showed that DEGs mainly enriched in multicellular organism development, detection of stimulus, diseases of signal transduction by growth factor receptors and second messengers, and olfactory signaling pathway. -

WO 2019/068007 Al Figure 2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/068007 Al 04 April 2019 (04.04.2019) W 1P O PCT (51) International Patent Classification: (72) Inventors; and C12N 15/10 (2006.01) C07K 16/28 (2006.01) (71) Applicants: GROSS, Gideon [EVIL]; IE-1-5 Address C12N 5/10 (2006.0 1) C12Q 1/6809 (20 18.0 1) M.P. Korazim, 1292200 Moshav Almagor (IL). GIBSON, C07K 14/705 (2006.01) A61P 35/00 (2006.01) Will [US/US]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., C07K 14/725 (2006.01) P.O. Box 4044, 7403635 Ness Ziona (TL). DAHARY, Dvir [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (21) International Application Number: Box 4044, 7403635 Ness Ziona (IL). BEIMAN, Merav PCT/US2018/053583 [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (22) International Filing Date: Box 4044, 7403635 Ness Ziona (E.). 28 September 2018 (28.09.2018) (74) Agent: MACDOUGALL, Christina, A. et al; Morgan, (25) Filing Language: English Lewis & Bockius LLP, One Market, Spear Tower, SanFran- cisco, CA 94105 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of national protection available): AE, AG, AL, AM, 62/564,454 28 September 2017 (28.09.2017) US AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/649,429 28 March 2018 (28.03.2018) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (71) Applicant: IMMP ACT-BIO LTD. -

Cancer Sequencing Service Data File Formats File Format V2.4 Software V2.4 December 2012
Cancer Sequencing Service Data File Formats File format v2.4 Software v2.4 December 2012 CGA Tools, cPAL, and DNB are trademarks of Complete Genomics, Inc. in the US and certain other countries. All other trademarks are the property of their respective owners. Disclaimer of Warranties. COMPLETE GENOMICS, INC. PROVIDES THESE DATA IN GOOD FAITH TO THE RECIPIENT “AS IS.” COMPLETE GENOMICS, INC. MAKES NO REPRESENTATION OR WARRANTY, EXPRESS OR IMPLIED, INCLUDING WITHOUT LIMITATION ANY IMPLIED WARRANTY OF MERCHANTABILITY OR FITNESS FOR A PARTICULAR PURPOSE OR USE, OR ANY OTHER STATUTORY WARRANTY. COMPLETE GENOMICS, INC. ASSUMES NO LEGAL LIABILITY OR RESPONSIBILITY FOR ANY PURPOSE FOR WHICH THE DATA ARE USED. Any permitted redistribution of the data should carry the Disclaimer of Warranties provided above. Data file formats are expected to evolve over time. Backward compatibility of any new file format is not guaranteed. Complete Genomics data is for Research Use Only and not for use in the treatment or diagnosis of any human subject. Information, descriptions and specifications in this publication are subject to change without notice. Copyright © 2011-2012 Complete Genomics Incorporated. All rights reserved. RM_DFFCS_2.4-01 Table of Contents Table of Contents Preface ...........................................................................................................................................................................................1 Conventions ................................................................................................................................................................................................. -
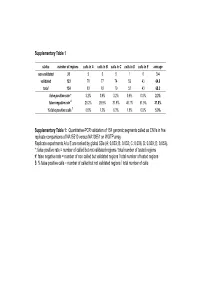
Quantitative-PCR Validation of 154 Genomic Segments Called As Cnvs in Five Replicat
Supplementary Table 1 status number of regions calls in A calls in B calls in C calls in D calls in E average non validated 31 5 6 5 1 0 3.4 validated 123 78 77 74 52 43 64.8 total 154 83 83 79 53 43 68.2 false positive rate * 3.2% 3.9% 3.2% 0.6% 0.0% 2.2% false negative rate # 29.2% 29.9% 31.8% 46.1% 51.9% 37.8% % false positive calls $ 6.0% 7.2% 6.3% 1.9% 0.0% 5.0% Supplementary Table 1: Quantitative-PCR validation of 154 genomic segments called as CNVs in five replicate comparisons of NA15510 versus NA10851 on WGTP array Replicate experiments A to E are ranked by global SDe (A: 0.033; B: 0.033; C: 0.036; D: 0.039; E: 0.053). *: false positive rate = number of called but not validated regions / total number of tested regions #: false negative rate = number of non called but validated regions / total number of tested regions $: % false positive calls = number of called but not validated regions / total number of calls False positive estimates for 500K EA CNV calls Total Rep1 Rep2 Rep3 Avg (unique) Validated 33 28 32 31 38 Not validated 2 2 2 2 5 Total 35 30 34 33 43 % False positive 5.71% 6.67% 5.88% 6.09% - % False negative 13.16% 26.32% 15.79% 18.42% - Supplementary Table 2A : Quantitative PCR validation of 43 unique CNV regions called as CNVs in three replicate comparisons of NA15510 versus NA10851 using the 500K EA array. -

Table S3. RAE Analysis of Well-Differentiated Liposarcoma
Table S3. RAE analysis of well-differentiated liposarcoma Model Chromosome Region start Region end Size q value freqX0* # genes Genes Amp 1 145009467 145122002 112536 0.097 21.8 2 PRKAB2,PDIA3P Amp 1 145224467 146188434 963968 0.029 23.6 10 CHD1L,BCL9,ACP6,GJA5,GJA8,GPR89B,GPR89C,PDZK1P1,RP11-94I2.2,NBPF11 Amp 1 147475854 148412469 936616 0.034 23.6 20 PPIAL4A,FCGR1A,HIST2H2BF,HIST2H3D,HIST2H2AA4,HIST2H2AA3,HIST2H3A,HIST2H3C,HIST2H4B,HIST2H4A,HIST2H2BE, HIST2H2AC,HIST2H2AB,BOLA1,SV2A,SF3B4,MTMR11,OTUD7B,VPS45,PLEKHO1 Amp 1 148582896 153398462 4815567 1.5E-05 49.1 152 PRPF3,RPRD2,TARS2,ECM1,ADAMTSL4,MCL1,ENSA,GOLPH3L,HORMAD1,CTSS,CTSK,ARNT,SETDB1,LASS2,ANXA9, FAM63A,PRUNE,BNIPL,C1orf56,CDC42SE1,MLLT11,GABPB2,SEMA6C,TNFAIP8L2,LYSMD1,SCNM1,TMOD4,VPS72, PIP5K1A,PSMD4,ZNF687,PI4KB,RFX5,SELENBP1,PSMB4,POGZ,CGN,TUFT1,SNX27,TNRC4,MRPL9,OAZ3,TDRKH,LINGO4, RORC,THEM5,THEM4,S100A10,S100A11,TCHHL1,TCHH,RPTN,HRNR,FLG,FLG2,CRNN,LCE5A,CRCT1,LCE3E,LCE3D,LCE3C,LCE3B, LCE3A,LCE2D,LCE2C,LCE2B,LCE2A,LCE4A,KPRP,LCE1F,LCE1E,LCE1D,LCE1C,LCE1B,LCE1A,SMCP,IVL,SPRR4,SPRR1A,SPRR3, SPRR1B,SPRR2D,SPRR2A,SPRR2B,SPRR2E,SPRR2F,SPRR2C,SPRR2G,LELP1,LOR,PGLYRP3,PGLYRP4,S100A9,S100A12,S100A8, S100A7A,S100A7L2,S100A7,S100A6,S100A5,S100A4,S100A3,S100A2,S100A16,S100A14,S100A13,S100A1,C1orf77,SNAPIN,ILF2, NPR1,INTS3,SLC27A3,GATAD2B,DENND4B,CRTC2,SLC39A1,CREB3L4,JTB,RAB13,RPS27,NUP210L,TPM3,C1orf189,C1orf43,UBAP2L,HAX1, AQP10,ATP8B2,IL6R,SHE,TDRD10,UBE2Q1,CHRNB2,ADAR,KCNN3,PMVK,PBXIP1,PYGO2,SHC1,CKS1B,FLAD1,LENEP,ZBTB7B,DCST2, DCST1,ADAM15,EFNA4,EFNA3,EFNA1,RAG1AP1,DPM3 Amp 1