The Evolution of a Bmp5 Enhancer in Primates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
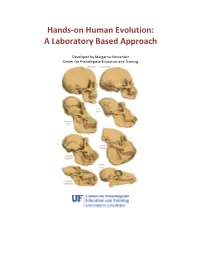
Hands-On Human Evolution: a Laboratory Based Approach
Hands-on Human Evolution: A Laboratory Based Approach Developed by Margarita Hernandez Center for Precollegiate Education and Training Author: Margarita Hernandez Curriculum Team: Julie Bokor, Sven Engling A huge thank you to….. Contents: 4. Author’s note 5. Introduction 6. Tips about the curriculum 8. Lesson Summaries 9. Lesson Sequencing Guide 10. Vocabulary 11. Next Generation Sunshine State Standards- Science 12. Background information 13. Lessons 122. Resources 123. Content Assessment 129. Content Area Expert Evaluation 131. Teacher Feedback Form 134. Student Feedback Form Lesson 1: Hominid Evolution Lab 19. Lesson 1 . Student Lab Pages . Student Lab Key . Human Evolution Phylogeny . Lab Station Numbers . Skeletal Pictures Lesson 2: Chromosomal Comparison Lab 48. Lesson 2 . Student Activity Pages . Student Lab Key Lesson 3: Naledi Jigsaw 77. Lesson 3 Author’s note Introduction Page The validity and importance of the theory of biological evolution runs strong throughout the topic of biology. Evolution serves as a foundation to many biological concepts by tying together the different tenants of biology, like ecology, anatomy, genetics, zoology, and taxonomy. It is for this reason that evolution plays a prominent role in the state and national standards and deserves thorough coverage in a classroom. A prime example of evolution can be seen in our own ancestral history, and this unit provides students with an excellent opportunity to consider the multiple lines of evidence that support hominid evolution. By allowing students the chance to uncover the supporting evidence for evolution themselves, they discover the ways the theory of evolution is supported by multiple sources. It is our hope that the opportunity to handle our ancestors’ bone casts and examine real molecular data, in an inquiry based environment, will pique the interest of students, ultimately leading them to conclude that the evidence they have gathered thoroughly supports the theory of evolution. -

Human Evolution: a Paleoanthropological Perspective - F.H
PHYSICAL (BIOLOGICAL) ANTHROPOLOGY - Human Evolution: A Paleoanthropological Perspective - F.H. Smith HUMAN EVOLUTION: A PALEOANTHROPOLOGICAL PERSPECTIVE F.H. Smith Department of Anthropology, Loyola University Chicago, USA Keywords: Human evolution, Miocene apes, Sahelanthropus, australopithecines, Australopithecus afarensis, cladogenesis, robust australopithecines, early Homo, Homo erectus, Homo heidelbergensis, Australopithecus africanus/Australopithecus garhi, mitochondrial DNA, homology, Neandertals, modern human origins, African Transitional Group. Contents 1. Introduction 2. Reconstructing Biological History: The Relationship of Humans and Apes 3. The Human Fossil Record: Basal Hominins 4. The Earliest Definite Hominins: The Australopithecines 5. Early Australopithecines as Primitive Humans 6. The Australopithecine Radiation 7. Origin and Evolution of the Genus Homo 8. Explaining Early Hominin Evolution: Controversy and the Documentation- Explanation Controversy 9. Early Homo erectus in East Africa and the Initial Radiation of Homo 10. After Homo erectus: The Middle Range of the Evolution of the Genus Homo 11. Neandertals and Late Archaics from Africa and Asia: The Hominin World before Modernity 12. The Origin of Modern Humans 13. Closing Perspective Glossary Bibliography Biographical Sketch Summary UNESCO – EOLSS The basic course of human biological history is well represented by the existing fossil record, although there is considerable debate on the details of that history. This review details both what is firmly understood (first echelon issues) and what is contentious concerning humanSAMPLE evolution. Most of the coCHAPTERSntention actually concerns the details (second echelon issues) of human evolution rather than the fundamental issues. For example, both anatomical and molecular evidence on living (extant) hominoids (apes and humans) suggests the close relationship of African great apes and humans (hominins). That relationship is demonstrated by the existing hominoid fossil record, including that of early hominins. -

Evolution of Grasping Among Anthropoids
doi: 10.1111/j.1420-9101.2008.01582.x Evolution of grasping among anthropoids E. POUYDEBAT,* M. LAURIN, P. GORCE* & V. BELSà *Handibio, Universite´ du Sud Toulon-Var, La Garde, France Comparative Osteohistology, UMR CNRS 7179, Universite´ Pierre et Marie Curie (Paris 6), Paris, France àUMR 7179, MNHN, Paris, France Keywords: Abstract behaviour; The prevailing hypothesis about grasping in primates stipulates an evolution grasping; from power towards precision grips in hominids. The evolution of grasping is hominids; far more complex, as shown by analysis of new morphometric and behavio- palaeobiology; ural data. The latter concern the modes of food grasping in 11 species (one phylogeny; platyrrhine, nine catarrhines and humans). We show that precision grip and precision grip; thumb-lateral behaviours are linked to carpus and thumb length, whereas primates; power grasping is linked to second and third digit length. No phylogenetic variance partitioning with PVR. signal was found in the behavioural characters when using squared-change parsimony and phylogenetic eigenvector regression, but such a signal was found in morphometric characters. Our findings shed new light on previously proposed models of the evolution of grasping. Inference models suggest that Australopithecus, Oreopithecus and Proconsul used a precision grip. very old behaviour, as it occurs in anurans, crocodilians, Introduction squamates and several therian mammals (Gray, 1997; Grasping behaviour is a key activity in primates to obtain Iwaniuk & Whishaw, 2000). On the contrary, the food. The hand is used in numerous activities of manip- precision grip, in which an object is held between the ulation and locomotion and is linked to several func- distal surfaces of the thumb and the index finger, is tional adaptations (Godinot & Beard, 1993; Begun et al., usually viewed as a derived function, linked to tool use 1997; Godinot et al., 1997). -

Reassessment of the Phylogenetic Relationships of the Late Miocene Apes Hispanopithecus and Rudapithecus Based on Vestibular Morphology
Reassessment of the phylogenetic relationships of the late Miocene apes Hispanopithecus and Rudapithecus based on vestibular morphology Alessandro Urciuolia,1, Clément Zanollib, Sergio Almécijaa,c,d, Amélie Beaudeta,e,f,g, Jean Dumoncelh, Naoki Morimotoi, Masato Nakatsukasai, Salvador Moyà-Solàa,j,k, David R. Begunl, and David M. Albaa,1 aInstitut Català de Paleontologia Miquel Crusafont, Universitat Autònoma de Barcelona, 08193 Barcelona, Spain; bUniv. Bordeaux, CNRS, MCC, PACEA, UMR 5199, F-33600 Pessac, France; cDivision of Anthropology, American Museum of Natural History, New York, NY 10024; dNew York Consortium in Evolutionary Primatology, New York, NY 10016; eDepartment of Archaeology, University of Cambridge, Cambridge CB2 1QH, United Kingdom; fSchool of Geography, Archaeology, and Environmental Studies, University of the Witwatersrand, Johannesburg, WITS 2050, South Africa; gDepartment of Anatomy, University of Pretoria, Pretoria 0001, South Africa; hLaboratoire Anthropology and Image Synthesis, UMR 5288 CNRS, Université de Toulouse, 31073 Toulouse, France; iLaboratory of Physical Anthropology, Graduate School of Science, Kyoto University, 606 8502 Kyoto, Japan; jInstitució Catalana de Recerca i Estudis Avançats, 08010 Barcelona, Spain; kUnitat d’Antropologia, Departament de Biologia Animal, Biologia Vegetal i Ecologia, Universitat Autònoma de Barcelona, 08193 Barcelona, Spain; and lDepartment of Anthropology, University of Toronto, Toronto, ON M5S 2S2, Canada Edited by Justin S. Sipla, University of Iowa, Iowa City, IA, and accepted by Editorial Board Member C. O. Lovejoy December 3, 2020 (received for review July 19, 2020) Late Miocene great apes are key to reconstructing the ancestral Dryopithecus and allied forms have long been debated. Until a morphotype from which earliest hominins evolved. Despite con- decade ago, several species of European apes from the middle sensus that the late Miocene dryopith great apes Hispanopithecus and late Miocene were included within this genus (9–16). -

A Unique Middle Miocene European Hominoid and the Origins of the Great Ape and Human Clade Salvador Moya` -Sola` A,1, David M
A unique Middle Miocene European hominoid and the origins of the great ape and human clade Salvador Moya` -Sola` a,1, David M. Albab,c, Sergio Alme´ cijac, Isaac Casanovas-Vilarc, Meike Ko¨ hlera, Soledad De Esteban-Trivignoc, Josep M. Roblesc,d, Jordi Galindoc, and Josep Fortunyc aInstitucio´Catalana de Recerca i Estudis Avanc¸ats at Institut Catala`de Paleontologia (ICP) and Unitat d’Antropologia Biolo`gica (Dipartimento de Biologia Animal, Biologia Vegetal, i Ecologia), Universitat Auto`noma de Barcelona, Edifici ICP, Campus de Bellaterra s/n, 08193 Cerdanyola del Valle`s, Barcelona, Spain; bDipartimento di Scienze della Terra, Universita`degli Studi di Firenze, Via G. La Pira 4, 50121 Florence, Italy; cInstitut Catala`de Paleontologia, Universitat Auto`noma de Barcelona, Edifici ICP, Campus de Bellaterra s/n, 08193 Cerdanyola del Valle`s, Barcelona, Spain; and dFOSSILIA Serveis Paleontolo`gics i Geolo`gics, S.L. c/ Jaume I nu´m 87, 1er 5a, 08470 Sant Celoni, Barcelona, Spain Edited by David Pilbeam, Harvard University, Cambridge, MA, and approved March 4, 2009 (received for review November 20, 2008) The great ape and human clade (Primates: Hominidae) currently sediments by the diggers and bulldozers. After 6 years of includes orangutans, gorillas, chimpanzees, bonobos, and humans. fieldwork, 150 fossiliferous localities have been sampled from the When, where, and from which taxon hominids evolved are among 300-m-thick local stratigraphic series of ACM, which spans an the most exciting questions yet to be resolved. Within the Afro- interval of 1 million years (Ϸ12.5–11.3 Ma, Late Aragonian, pithecidae, the Kenyapithecinae (Kenyapithecini ؉ Equatorini) Middle Miocene). -

Class 2: Early Hominids
Earliest Hominins CHARLES J VELLA, PHD JULY 25 2018 This is latest theory of how Lucy died! We are Mammals 3 defining mammalian traits: hair, mammary glands, homeothermy Mammalian traits show an adaptation for adaptability Miocene era: 23 to 5 Ma, Warmer global period • Ape grade: Planet of the apes • Over 30 genera and 100 species of ape – compared with 6 today • Location: Africa and Eurasia Proconsul: 25 to 23 Ma, during Miocene; arboreal quadruped Primates • Larger body size • Larger brain • Complete stereoscopic vision • Longer gestation, infancy, life span • More k-selected (tend towards single offspring) • Greater dependency on learned behavior • More social Great Apes Bonobos and Chimps split ~1 Ma Superfamily: Hominoidea Gibbons, Gorillas, Orangutan, Chimpanzee, Human Greater encephalization (brain to body ratio) = smarter larger body, brachiation, social complexity, lack of tail Why did Newt Gingrich recommend this book to all new politicians? Detailed and thoroughly engrossing account of ape rivalries and coalitions. Machiavellianism: political behavior is rooted at a level of development that is below the cognitive and is as much instinctive as it is learned. de Waal 1982 De Waal: Machiavellian IQ Machiavelli's The Prince: Frans de Waal introduced the term 'Machiavellian Intelligence' to describe the social and political behavior of chimpanzees Social behaviors: reconciliation, alliance, and sabotage Tactical deception in primates: Vervet monkeys use false predator alarm calls to get extra food Chimpanzees use deception to mate with females belonging to alpha male Chimpanzee cultures • Chimp Cultures: shared behaviors in different communities: • pounding actions • fishing; • probing; • forcing • comfort behavior • miscellaneous exploitation of vegetation properties • exploitation of leaf properties; • grooming; • attention-getting. -

Cognitive Inferences in Fossil Apes (Primates, Hominoidea): Does Encephalization Reflect Intelligence?
JASs Invited Reviews Journal of Anthropological Sciences Vol. 88 (2010), pp. 11-48 Cognitive inferences in fossil apes (Primates, Hominoidea): does encephalization reflect intelligence? David M. Alba Institut Català de Paleontologia, Universitat Autònoma de Barcelona. Edifici ICP, Campus de la UAB s/n, 08193 Cerdanyola del Vallès, Barcelona (Spain); Dipartimento di Scienze della Terra, Università degli Studi di Firenze. Via G. La Pira 4, 50121 Firenze (Italy); e-mail: [email protected] Summary – Paleobiological inferences on general cognitive abilities (intelligence) in fossil hominoids strongly rely on relative brain size or encephalization, computed by means of allometric residuals, quotients or constants. This has been criticized on the basis that it presumably fails to reflect the higher intelligence of great apes, and absolute brain size has been favored instead. Many problems of encephalization metrics stem from the decrease of allometric slopes towards lower taxonomic level, thus making it difficult to determine at what level encephalization metrics have biological meaning. Here, the hypothesis that encephalization can be used as a good neuroanatomical proxy for intelligence is tested at two different taxonomic levels. A significant correlation is found between intelligence and encephalization only at a lower taxonomic level, i.e. on the basis of a low allometric slope, irrespective of whether species data or independent contrasts are employed. This indicates that higher-level slopes, resulting from encephalization grade shifts between subgroups (including hylobatids vs. great apes), do not reflect functional equivalence, whereas lower-level metrics can be employed as a paleobiological proxy for intelligence. Thus, in accordance to intelligence rankings, lower-level metrics indicate that great apes are more encephalized than both monkeys and hylobatids. -

Explore Your Inner Animals Worksheet
TEACHER MATERIALS EXPLORE YOUR INNER ANIMALS WORKSHEET OVERVIEW This worksheet accompanies the Click & Learn “Explore Your Inner Animals” (http://www.hhmi.org/biointeractive/explore- your-inner-animals). As they work through the Click & Learn, students explore multiple lines of evidence for common descent found among the bodies and cells of both extinct and extant organisms. This content was also featured in the film Your Inner Fish and in the Great Transitions: The Origin of Tetrapods short film. Visit http://www.hhmi.org/biointeractive/your-inner-fish for more information. The student worksheet is designed to help ensure students successfully navigate the interactive and can be completed in class or assigned as homework. Students may complete all of the different sections or only some of them. KEY CONCEPTS AND LESSON OBJECTIVES • Species descend from other species. Even distantly related species, like humans and sponges, can trace their shared ancestry back to a common ancestor. • Evidence for common descent includes the fossil record and anatomical, genetic, and developmental homologies among organisms. • The fossil record provides a history of life on Earth. It includes fossils with features that are intermediate, or transitional, between those of major groups of animals. • When a series of transitional fossils are viewed together, they reveal the gradual sequence of change connecting one major group to another. • An organism’s DNA codes for proteins that result in an organism’s visible traits. • Scientists infer function and behavior from anatomical structures. • Natural selection is the process by which heritable traits that confer a survival and/or reproductive advantage to individuals that possess them increase in frequency within a population over generations. -

Genetics and Genomics in the Study of Human Evolution and Prehistory
Paweł Golik Instytut Genetyki i Biotechnologii Wydział Biologii, Uniwersytet Warszawski Genetics and genomics in the study of human evolution and prehistory The presentation will be available How do we know about the past? • Fossil remains - paleontology • Most of evolutionary history • Fragmentary, require favorable conditions to be preserved © nationalgeographic.com DNA as the source of information • Comparative analysis of contemporary sequences • phylogenetic inference • population analysis (admixture, coalescence) DNA as the source of information • Ancient DNA (aDNA) • partial and complete sequences • limits in hominids: ~400 kYA (thousands of years) (partial, mtDNA) • oldest complete hominin genome ~ 120 kYA (neanderthal) • oldest H. sapiens genome - 45 kYA (Ust’- Ishim, Siberia) • oldest complete genome - > 700 kYA years (a horse species) - preserved in permafrost Bence Viola, MPI EVA, nature.com aDNA Hofreiter et al. 2014, Bioessays 36 Ancestors? Multiple hominid fossils found. The relations are uncertain: ancestors or cousins? The tree • Evolution is not linear • The status of many fossils often unclear - cannot be assigned to a particular branch • There is no “missing link” - current paleontology has many fossils of transitory forms from ape-like to human-like We are animals! Linnaeus, 1735 Systema Naturae We are primates Humans Chimps Gorilla Orangutan milionów lat temu Macaque Gibbon MY (millions of years) Taxonomy • Hominids - the family Hominidae: • currently: humans, chimpanzees (and bonobos), gorillas, orangutans • oldest -

The Life History of Ardipithecus Ramidus: a Heterochronic Model of Sexual and Social Maturation
Early origins of human sexual and social maturation Gary Clark, Maciej Henneberg ANTHROPOLOGICAL REVIEW • Vol. 78 (2), 109–132 (2015) The life history of Ardipithecus ramidus: a heterochronic model of sexual and social maturation Gary Clark, Maciej Henneberg Biological Anthropology and Comparative Anatomy Unit University of Adelaide Medical School, Australia ABSTRACT: In this paper we analyse the ontogeny of craniofacial growth in Ardipithecus ramidus in the context of its possible social and environmental determinants. We sought to test the hypothesis that this form of early hominin evolved a specific adult craniofacial morphology via heterochronic dissociation of growth trajectories. We suggest the lack of sexual dimorphism in craniofacial morphology provides evidence for a suite of adult behavioral adaptations, and consequently an ontogeny, unlike any other species of extant ape. The lack of sexually dimorphic craniofacial morphology suggests A. ramidus males adopted reproductive strategies that did not require male on male conflict. Male investment in the maternal metabolic budget and/or paternal investment in offspring may have been reproductive strategies adopted by males. Such strategies would account for the absence of innate morphological armoury in males. Consequently, A. rami- dus would have most likely had sub-adult periods of socialisation unlike that of any extant ape. We also argue that A.ramidus and chimpanzee craniofacial morphology are apomorphic, each representing a derived condition relative to that of the common ancestor, with A. ramidus developing its orthognatic condition via paedomoporhosis, and chimpanzees evolving increased prognathism via peramorphosis. In contrast we suggest cranial volume and life history trajectories may be synapomorphic traits that both species inher- ited and retained form a putative common ancestral condition. -

Primates, Mammalia) from Anatolia
A NEW SPECIES OF TORTONIAN ANTHROPOID (PRIMATES, MAMMALIA) FROM ANATOLIA İbrahim TEKKAYA Mineral Research and Exploration Institute of Turkey SUMMARY. — We discovered in 1973—during a paleontological research in the continental Series of the Middle Miocene of Turkey—a fossil mandible of an Anthropoid which is significant for Turkey and Eurasia. The mandible was found at Hırsızderesi, near the town of Çandır (Kalecik), in Ankara Province. We examined the specimen and consider that from the anthropological point of view it has both Anthropoid and Homo characteristics. Since both primitive and advanced characteristics were observed in this specimen, we assigned this mandible to a new species of Sivapithecııs genera and called it Sivapithecus alpani.1 INTRODUCTION The Mammalian biozone of Çandır is one of the most important discoveries resulting from the paleontological investigations of the last years. For this reason it is hoped that the representatives of the continental Mammalian fauna in this Series will solve many problems regarding the Cenozoic of Turkey and the migration of Mammalian fauna from Asia to Europe. This Mammalian fauna was first discovered in 1968 by the Turkish-German teams of investi- gators. That year a small excavation was made., and again in 1969 the same group made a second small excavation at the same locality. The last investigation was made in 1973, in the area between Kale- cik-Çandır-Çankırı by the Turkish Vertebrate Paleontology Group of the M.T.A. Institute. This most recent work uncovered valuable evidence of Mammalian fossils which were studied and are now exhibited in the Natural History Museum, M.T.A. -

FYI's Compiled.Cwk
3.987 - Human Origins and Evolution Spring 2006 List of FYI’s - e-mailed “handouts” and communications The following items were circulated to students before or after lectures as background and/or follow-up information on questions and points raised in specific lectures. Lecture 6 FYI #1 Oreopithecus Lecture 8 FYI #2 Tooth formation rates Lecture 11 FYI #4 The gluteus maximus Lecture 8 FYI #5 On the diet of australopithecines Lecture 12 FYI #6 On early stone tool use Lecture 17 FYI #7 Headstrong hominids Lecture 17 FYI #9 Tan-Tan Acheulian figurine Lecture 17 FYI #10 Schoningen spear Lecture 15 FYI #11 The role of cooking in human evolution Lecture 21 FYI #12 Early sapiens mtDNA in Australia Lecture 21 FYI #13 Possible Neandertal/sapiens hybrid? Lecture 20 FYI #14 Neandertal mobility patterns Lecture 22 FYI #17 One last mystery at Dolni Vestonice -------------------------- FYI #1 - Oreopithecus Date: 3/2/06 I threatened you with some spam about Oreopithecus, a Miocene, possibly bipedal, ape. Well here it is. As I mentioned one of the really interesting recent discoveries is that there may have been another genus of now extinct apes which independently developed bipedalism. The candidate is Oreopithecus a 9 to 7 million year old genus of European ape, however, your textbook authors don't provide any real discussion of this evidence or this possibility. The genus Oreopithecus has been recognized for over 130 years, it being one of the first fossil apes to be discovered and described. Below are several references and abstracts of relatively recent papers which have reanalyzed the Oreopithecus remains and have somewhat controversially proposed that Oreopithecus was a habitual biped.