RLIP76: a Structural and Functional Triumvirate
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Localization of Ralb Signaling at Endomembrane Compartments and Its Modulation by Autophagy Received: 14 January 2019 Manish Kumar Singh1,2, Alexandre P
www.nature.com/scientificreports Corrected: Publisher Correction OPEN Localization of RalB signaling at endomembrane compartments and its modulation by autophagy Received: 14 January 2019 Manish Kumar Singh1,2, Alexandre P. J. Martin1,2, Carine Jofre 3, Giulia Zago1,2, Accepted: 30 May 2019 Jacques Camonis1,2, Mathieu Coppey1,4 & Maria Carla Parrini1,2 Published online: 20 June 2019 The monomeric GTPase RalB controls crucial physiological processes, including autophagy and invasion, but it still remains unclear how this multi-functionality is achieved. Previously, we reported that the RalGEF (Guanine nucleotide Exchange Factor) RGL2 binds and activates RalB to promote invasion. Here we show that RGL2, a major activator of RalB, is also required for autophagy. Using a novel automated image analysis method, Endomapper, we quantifed the endogenous localization of the RGL2 activator and its substrate RalB at diferent endomembrane compartments, in an isogenic normal and Ras-transformed cell model. In both normal and Ras-transformed cells, we observed that RGL2 and RalB substantially localize at early and recycling endosomes, and to lesser extent at autophagosomes, but not at trans-Golgi. Interestingly the use of a FRET-based RalB biosensor indicated that RalB signaling is active at these endomembrane compartments at basal level in rich medium. Furthermore, induction of autophagy by nutrient starvation led to a considerable reduction of early and recycling endosomes, in contrast to the expected increase of autophagosomes, in both normal and Ras-transformed cells. However, autophagy mildly afected relative abundances of both RGL2 and RalB at early and recycling endosomes, and at autophagosomes. Interestingly, RalB activity increased at autophagosomes upon starvation in normal cells. -
![Downloaded from [266]](https://docslib.b-cdn.net/cover/7352/downloaded-from-266-347352.webp)
Downloaded from [266]
Patterns of DNA methylation on the human X chromosome and use in analyzing X-chromosome inactivation by Allison Marie Cotton B.Sc., The University of Guelph, 2005 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in The Faculty of Graduate Studies (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2012 © Allison Marie Cotton, 2012 Abstract The process of X-chromosome inactivation achieves dosage compensation between mammalian males and females. In females one X chromosome is transcriptionally silenced through a variety of epigenetic modifications including DNA methylation. Most X-linked genes are subject to X-chromosome inactivation and only expressed from the active X chromosome. On the inactive X chromosome, the CpG island promoters of genes subject to X-chromosome inactivation are methylated in their promoter regions, while genes which escape from X- chromosome inactivation have unmethylated CpG island promoters on both the active and inactive X chromosomes. The first objective of this thesis was to determine if the DNA methylation of CpG island promoters could be used to accurately predict X chromosome inactivation status. The second objective was to use DNA methylation to predict X-chromosome inactivation status in a variety of tissues. A comparison of blood, muscle, kidney and neural tissues revealed tissue-specific X-chromosome inactivation, in which 12% of genes escaped from X-chromosome inactivation in some, but not all, tissues. X-linked DNA methylation analysis of placental tissues predicted four times higher escape from X-chromosome inactivation than in any other tissue. Despite the hypomethylation of repetitive elements on both the X chromosome and the autosomes, no changes were detected in the frequency or intensity of placental Cot-1 holes. -

The Role of Rala and Ralb in Cancer Samuel C
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School 4-7-2008 The Role of RalA and RalB in Cancer Samuel C. Falsetti University of South Florida Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the American Studies Commons Scholar Commons Citation Falsetti, Samuel C., "The Role of RalA and RalB in Cancer" (2008). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/232 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. The Role of RalA and RalB in Cancer By Samuel C. Falsetti A dissertation submitted in partial fulfillment Of the requirements for the degree of Doctor of Philosophy Department of Molecular Medicine College of Medicine University of South Florida Major Professor: Saïd M. Sebti, Ph.D. Larry P. Solomonson, Ph.D. Gloria C. Ferreira, Ph.D. Srikumar Chellapan, Ph.D. Gary Reuther, Ph.D. Douglas Cress, Ph.D. Date of Approval: April 7th, 2008 Keywords: Ras, RACK1, Geranylgeranyltransferase I inhibitors, ovarian cancer, proteomics © Copyright 2008, Samuel C. Falsetti Dedication This thesis is dedicated to my greatest supporter, my wife. Without her loving advice and patience none of this would be possible. Acknowledgments I would like to extend my sincere gratitude to my wife, Nicole. She is the most inspirational person in my life and I am honored to be with her; in truth, this degree ought to come with two names printed on it. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -
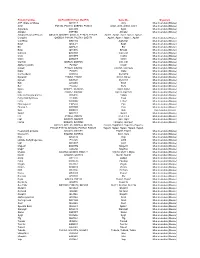
Gene Ids Organism ATP Citrate Synthase Q3V117 Acly
Protein Families UniProtKB ID (from MudPIT) Gene IDs Organism ATP citrate synthase Q3V117 Acly Mus musculus (Mouse) Actin P68134, P60710, Q8BFZ3, P68033 Acta1, Actb, Actbl2, Actc1 Mus musculus (Mouse) Argonaute Q8CJG0 Ago2 Mus musculus (Mouse) Ahnak2 E9PYB0 Ahnak2 Mus musculus (Mouse) Adaptor Related Protein Q8CC13, Q8CBB7, Q3UHJ0, P17426, P17427, Ap1b1, Ap1g1, Aak1, Ap2a1, Ap2a2, Complex Q9DBG3, P84091, P62743, Q9Z1T1 Ap2b1, Ap2m1, Ap2s1 , Ap3b1 Mus musculus (Mouse) V-ATPase Q9Z1G4 Atp6v0a1 Mus musculus (Mouse) Bag3 Q9JLV1 Bag3 Mus musculus (Mouse) Bcr Q6PAJ1 Bcr Mus musculus (Mouse) Bmp Q91Z96 Bmp2k Mus musculus (Mouse) Calcoco Q8CGU1 Calcoco1 Mus musculus (Mouse) Ccdc D3YZP9 Ccdc6 Mus musculus (Mouse) Clint1 Q5SUH7 Clint1 Mus musculus (Mouse) Clathrin Q6IRU5, Q68FD5 Cltb, Cltc Mus musculus (Mouse) Alpha-crystallin P23927 Cryab Mus musculus (Mouse) Casein P19228, Q02862 Csn1s1, Csn1s2a Mus musculus (Mouse) Dab2 P98078 Dab2 Mus musculus (Mouse) Connecdenn Q8K382 Dennd1a Mus musculus (Mouse) Dynamin P39053, P39054 Dnm1, Dnm2 Mus musculus (Mouse) Dynein Q9JHU4 Dync1h1 Mus musculus (Mouse) Edc Q3UJB9 Edc4 Mus musculus (Mouse) Eef P58252 Eef2 Mus musculus (Mouse) Epsin Q80VP1, Q5NCM5 Epn1, Epn2 Mus musculus (Mouse) Eps P42567, Q60902 Eps15, Eps15l1 Mus musculus (Mouse) Fatty acid binding protein Q05816 Fabp5 Mus musculus (Mouse) Fatty Acid Synthase P19096 Fasn Mus musculus (Mouse) Fcho Q3UQN2 Fcho2 Mus musculus (Mouse) Fibrinogen A E9PV24 Fga Mus musculus (Mouse) Filamin A Q8BTM8 Flna Mus musculus (Mouse) Gak Q99KY4 Gak Mus musculus (Mouse) -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

Phosphorylation of Ralb Is Important for Bladder Cancer Cell Growth and Metastasis
Published OnlineFirst October 12, 2010; DOI: 10.1158/0008-5472.CAN-10-0952 Published OnlineFirst on October 12, 2010 as 10.1158/0008-5472.CAN-10-0952 Therapeutics, Targets, and Chemical Biology Cancer Research Phosphorylation of RalB Is Important for Bladder Cancer Cell Growth and Metastasis Hong Wang1, Charles Owens1, Nidhi Chandra1, Mark R. Conaway2, David L. Brautigan3,4, and Dan Theodorescu5 Abstract RalA and RalB are monomeric G proteins that are 83% identical in amino acid sequence but have paralogue- specific effects on cell proliferation, metastasis, and apoptosis. Using in vitro kinase assays and phosphosite- specific antibodies, here we show phosphorylation of RalB by protein kinase C (PKC) and RalA by protein kinase A. We used mass spectrometry and site-directed mutagenesis to identify S198 as the primary PKC phosphorylation site in RalB. Phorbol ester [phorbol 12-myristate 13-acetate (PMA)] treatment of human blad- der carcinoma cells induced S198 phosphorylation of stably expressed FLAG-RalB as well as endogenous RalB. PMA treatment caused RalB translocation from the plasma membrane to perinuclear regions in a S198 phos- phorylation–dependent manner. Using RNA interference depletion of RalB followed by rescue with wild-type RalB or RalB(S198A) as well as overexpression of wild-type RalB or RalB(S198A) with and without PMA stimulation, we show that phosphorylation of RalB at S198 is necessary for actin cytoskeletal organization, anchorage-independent growth, cell migration, and experimental lung metastasis of T24 or UMUC3 human bladder cancer cells. In addition, UMUC3 cells transfected with a constitutively active RalB(G23V) exhibited enhanced subcutaneous tumor growth, whereas those transfected with phospho-deficient RalB(G23V-S198A) were indistinguishable from control cells. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Human RALBP1 Peptide (DAG-P1058) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Human RALBP1 peptide (DAG-P1058) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Antigen Description RALBP1 plays a role in receptor-mediated endocytosis and is a downstream effector of the small GTP-binding protein RAL (see RALA; MIM 179550). Small G proteins, such as RAL, have GDP- bound inactive and GTP-bound active forms, which shift from the inactive to the active state through the action of RALGDS (MIM 601619), which in turn is activated by RAS (see HRAS; MIM 190020) (summary by Feig, 2003 [PubMed 12888294]).[supplied by OMIM, Nov 2010] Specificity Expressed ubiquitously but at low levels. Shows a strong expression in the erythrocytes. Nature Synthetic Expression System N/A Conjugate Unconjugated Sequence Similarities Contains 1 Rho-GAP domain. Cellular Localization Membrane. Procedure None Format Liquid Preservative None Storage Shipped at 4°C. Upon delivery aliquot and store at -20°C or -80°C. Avoid repeated freeze / thaw cycles. Information available upon request. ANTIGEN GENE INFORMATION Gene Name RALBP1 ralA binding protein 1 [ Homo sapiens (human) ] Official Symbol RALBP1 Synonyms RALBP1; ralA binding protein 1; RIP1; RLIP1; RLIP76; ralA-binding protein 1; DNP-SG ATPase; ral-interacting protein 1; 76 kDa Ral-interacting protein; dinitrophenyl S-glutathione ATPase; Entrez Gene ID 10928 mRNA Refseq NM_006788.3 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved Protein -

Characterization of the Macrophage Transcriptome in Glomerulonephritis-Susceptible and -Resistant Rat Strains
Genes and Immunity (2011) 12, 78–89 & 2011 Macmillan Publishers Limited All rights reserved 1466-4879/11 www.nature.com/gene ORIGINAL ARTICLE Characterization of the macrophage transcriptome in glomerulonephritis-susceptible and -resistant rat strains K Maratou1, J Behmoaras2, C Fewings1, P Srivastava1, Z D’Souza1, J Smith3, L Game4, T Cook2 and T Aitman1 1Physiological Genomics and Medicine Group, MRC Clinical Sciences Centre, Imperial College London, London, UK; 2Centre for Complement and Inflammation Research, Imperial College London, London, UK; 3Renal Section, Imperial College London, London, UK and 4Genomics Laboratory, MRC Clinical Sciences Centre, London, UK Crescentic glomerulonephritis (CRGN) is a major cause of rapidly progressive renal failure for which the underlying genetic basis is unknown. Wistar–Kyoto (WKY) rats show marked susceptibility to CRGN, whereas Lewis rats are resistant. Glomerular injury and crescent formation are macrophage dependent and mainly explained by seven quantitative trait loci (Crgn1–7). Here, we used microarray analysis in basal and lipopolysaccharide (LPS)-stimulated macrophages to identify genes that reside on pathways predisposing WKY rats to CRGN. We detected 97 novel positional candidates for the uncharacterized Crgn3–7. We identified 10 additional secondary effector genes with profound differences in expression between the two strains (45-fold change, o1% false discovery rate) for basal and LPS-stimulated macrophages. Moreover, we identified eight genes with differentially expressed alternatively spliced isoforms, by using an in-depth analysis at the probe level that allowed us to discard false positives owing to polymorphisms between the two rat strains. Pathway analysis identified several common linked pathways, enriched for differentially expressed genes, which affect macrophage activation. -

The Small G-Protein Rala Promotes Progression and Metastasis of Triple- Negative Breast Cancer Katie A
Thies et al. Breast Cancer Research (2021) 23:65 https://doi.org/10.1186/s13058-021-01438-3 RESEARCH ARTICLE Open Access The small G-protein RalA promotes progression and metastasis of triple- negative breast cancer Katie A. Thies1,2, Matthew W. Cole1,2, Rachel E. Schafer1,2, Jonathan M. Spehar1,2, Dillon S. Richardson1,2, Sarah A. Steck1,2, Manjusri Das1,2, Arthur W. Lian1,2, Alo Ray1,2, Reena Shakya1,3, Sue E. Knoblaugh4, Cynthia D. Timmers5,6, Michael C. Ostrowski5,7, Arnab Chakravarti1,2, Gina M. Sizemore1,2 and Steven T. Sizemore1,2* Abstract Background: Breast cancer (BC) is the most common cancer in women and the leading cause of cancer-associated mortality in women. In particular, triple-negative BC (TNBC) has the highest rate of mortality due in large part to the lack of targeted treatment options for this subtype. Thus, there is an urgent need to identify new molecular targets for TNBC treatment. RALA and RALB are small GTPases implicated in growth and metastasis of a variety of cancers, although little is known of their roles in BC. Methods: The necessity of RALA and RALB for TNBC tumor growth and metastasis were evaluated in vivo using orthotopic and tail-vein models. In vitro, 2D and 3D cell culture methods were used to evaluate the contributions of RALA and RALB during TNBC cell migration, invasion, and viability. The association between TNBC patient outcome and RALA and RALB expression was examined using publicly available gene expression data and patient tissue microarrays. Finally, small molecule inhibition of RALA and RALB was evaluated as a potential treatment strategy for TNBC in cell line and patient-derived xenograft (PDX) models.