Interactome of Mirnas and Transcriptome of Human Umbilical Cord Endothelial Cells Exposed to Short-Term Simulated Microgravity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

I ABSTRACT KHAREL, PRAKASH, Ph.D., December 2018 Chemistry
ABSTRACT KHAREL, PRAKASH, Ph.D., December 2018 Chemistry & Biochemistry RNA OXIDATION IN MULTIPLE SCLEROSIS NEURODEGENERATION (204 PP.) Dissertation advisor: Soumitra Basu An increase in the production of reactive oxygen species (ROS) and the inability of cellular machinery to adequately neutralize the ROS thus produced, lead the cells towards oxidative stress. ROS can damage all major classes of biomolecules including proteins, DNA and RNA. Recent findings show the evidence of high level of RNA oxidation in the neuronal cells of many neurological disorders suggesting a link between RNA oxidation and neurodegeneration. We have recently discovered the presence of extensive oxidative damage in the RNA molecules of neuronal cells in postmortem brains of multiple sclerosis (MS) patients. Working on the MS model system, we have established the identity of selectively oxidized mRNA molecules under MS microenvironment in human neuronal cells. Our study reveals that many of the mRNAs linked to various neurological pathways are selectively oxidized under oxidative stress. We have demonstrated the functional consequences of mRNA oxidation in two neuropathology related mRNAs (namely Nat8l and Nlrp3 mRNA). N-acetyl aspartate transferase 8 like protein (NAT8L) is a key trans-mitochondrial membrane protein that catalyzes the transfer of acetyl group from acetyl-CoA to aspartate to form N-acetyl aspartate (NAA) and transports NAA to neuronal cytoplasm. We have discovered that the oxidation in Nat8l mRNA molecule i results in the reduced expression of NAT8L enzyme. We also observed a reduced level of NAA present in the neuronal cells under oxidative stress. Using the neuronal tissue culture and MS animal model (cuprizone mouse model), we have established that a higher mRNA oxidation results in a reduced expression of NAT8L protein, which presumably contributes to the MS disease progression by weakening the myelin production machinery. -
![Downloaded from [266]](https://docslib.b-cdn.net/cover/7352/downloaded-from-266-347352.webp)
Downloaded from [266]
Patterns of DNA methylation on the human X chromosome and use in analyzing X-chromosome inactivation by Allison Marie Cotton B.Sc., The University of Guelph, 2005 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in The Faculty of Graduate Studies (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2012 © Allison Marie Cotton, 2012 Abstract The process of X-chromosome inactivation achieves dosage compensation between mammalian males and females. In females one X chromosome is transcriptionally silenced through a variety of epigenetic modifications including DNA methylation. Most X-linked genes are subject to X-chromosome inactivation and only expressed from the active X chromosome. On the inactive X chromosome, the CpG island promoters of genes subject to X-chromosome inactivation are methylated in their promoter regions, while genes which escape from X- chromosome inactivation have unmethylated CpG island promoters on both the active and inactive X chromosomes. The first objective of this thesis was to determine if the DNA methylation of CpG island promoters could be used to accurately predict X chromosome inactivation status. The second objective was to use DNA methylation to predict X-chromosome inactivation status in a variety of tissues. A comparison of blood, muscle, kidney and neural tissues revealed tissue-specific X-chromosome inactivation, in which 12% of genes escaped from X-chromosome inactivation in some, but not all, tissues. X-linked DNA methylation analysis of placental tissues predicted four times higher escape from X-chromosome inactivation than in any other tissue. Despite the hypomethylation of repetitive elements on both the X chromosome and the autosomes, no changes were detected in the frequency or intensity of placental Cot-1 holes. -

The Expression of Non-Coding Rnas and Their Target Molecules in Rheumatoid Arthritis
International Journal of Molecular Sciences Review The Expression of Non-Coding RNAs and Their Target Molecules in Rheumatoid Arthritis: A Molecular Basis for Rheumatoid Pathogenesis and Its Potential Clinical Applications Chang-Youh Tsai 1,*,† , Song-Chou Hsieh 2,†, Chih-Wei Liu 1, Cheng-Hsun Lu 2,3 , Hsien-Tzung Liao 1, Ming-Han Chen 1, Ko-Jen Li 2, Cheng-Han Wu 2,3, Cheih-Yu Shen 2,3 , Yu-Min Kuo 2,3 and Chia-Li Yu 2,* 1 Division of Allergy, Immunology & Rheumatology, Taipei Veterans General Hospital, National Yang-Ming Chiao-Tung University, Taipei 11217, Taiwan; [email protected] (C.-W.L.); [email protected] (H.-T.L.); [email protected] (M.-H.C.) 2 Department of Internal Medicine, National Taiwan University Hospital, National Taiwan University College of Medicine, Taipei 10002, Taiwan; [email protected] (S.-C.H.); [email protected] (C.-H.L.); [email protected] (K.-J.L.); [email protected] (C.-H.W.); [email protected] (C.-Y.S.); [email protected] (Y.-M.K.) 3 Institute of Molecular Medicine, National Taiwan University College of Medicine, Taipei 10002, Taiwan * Correspondence: [email protected] (C.-Y.T.); [email protected] (C.-L.Y.) † The two co-first authors contributed equally in the preparation of the manuscript. Abstract: Rheumatoid arthritis (RA) is a typical autoimmune-mediated rheumatic disease presenting Citation: Tsai, C.-Y.; Hsieh, S.-C.; Liu, C.-W.; Lu, C.-H.; Liao, H.-T.; Chen, as a chronic synovitis in the joint. The chronic synovial inflammation is characterized by hyper- M.-H.; Li, K.-J.; Wu, C.-H.; Shen, C.-Y.; vascularity and extravasation of various immune-related cells to form lymphoid aggregates where an Kuo, Y.-M.; et al. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -
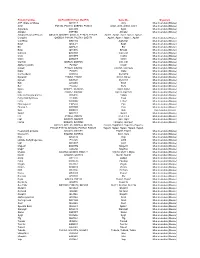
Gene Ids Organism ATP Citrate Synthase Q3V117 Acly
Protein Families UniProtKB ID (from MudPIT) Gene IDs Organism ATP citrate synthase Q3V117 Acly Mus musculus (Mouse) Actin P68134, P60710, Q8BFZ3, P68033 Acta1, Actb, Actbl2, Actc1 Mus musculus (Mouse) Argonaute Q8CJG0 Ago2 Mus musculus (Mouse) Ahnak2 E9PYB0 Ahnak2 Mus musculus (Mouse) Adaptor Related Protein Q8CC13, Q8CBB7, Q3UHJ0, P17426, P17427, Ap1b1, Ap1g1, Aak1, Ap2a1, Ap2a2, Complex Q9DBG3, P84091, P62743, Q9Z1T1 Ap2b1, Ap2m1, Ap2s1 , Ap3b1 Mus musculus (Mouse) V-ATPase Q9Z1G4 Atp6v0a1 Mus musculus (Mouse) Bag3 Q9JLV1 Bag3 Mus musculus (Mouse) Bcr Q6PAJ1 Bcr Mus musculus (Mouse) Bmp Q91Z96 Bmp2k Mus musculus (Mouse) Calcoco Q8CGU1 Calcoco1 Mus musculus (Mouse) Ccdc D3YZP9 Ccdc6 Mus musculus (Mouse) Clint1 Q5SUH7 Clint1 Mus musculus (Mouse) Clathrin Q6IRU5, Q68FD5 Cltb, Cltc Mus musculus (Mouse) Alpha-crystallin P23927 Cryab Mus musculus (Mouse) Casein P19228, Q02862 Csn1s1, Csn1s2a Mus musculus (Mouse) Dab2 P98078 Dab2 Mus musculus (Mouse) Connecdenn Q8K382 Dennd1a Mus musculus (Mouse) Dynamin P39053, P39054 Dnm1, Dnm2 Mus musculus (Mouse) Dynein Q9JHU4 Dync1h1 Mus musculus (Mouse) Edc Q3UJB9 Edc4 Mus musculus (Mouse) Eef P58252 Eef2 Mus musculus (Mouse) Epsin Q80VP1, Q5NCM5 Epn1, Epn2 Mus musculus (Mouse) Eps P42567, Q60902 Eps15, Eps15l1 Mus musculus (Mouse) Fatty acid binding protein Q05816 Fabp5 Mus musculus (Mouse) Fatty Acid Synthase P19096 Fasn Mus musculus (Mouse) Fcho Q3UQN2 Fcho2 Mus musculus (Mouse) Fibrinogen A E9PV24 Fga Mus musculus (Mouse) Filamin A Q8BTM8 Flna Mus musculus (Mouse) Gak Q99KY4 Gak Mus musculus (Mouse) -

Micrornas As New Regulators of Neutrophil Extracellular Trap Formation
International Journal of Molecular Sciences Review MicroRNAs as New Regulators of Neutrophil Extracellular Trap Formation Sonia Águila † , Ascensión M. de los Reyes-García †, María P. Fernández-Pérez, Laura Reguilón-Gallego, Laura Zapata-Martínez, Inmaculada Ruiz-Lorente, Vicente Vicente , Rocío González-Conejero *,‡ and Constantino Martínez *,‡ Department of Hematology and Medical Oncology, Morales Meseguer University Hospital, Centro Regional de Hemodonación, Universidad de Murcia, IMIB, C/Ronda de Garay S/N, 30003 Murcia, Spain; [email protected] (S.Á.); [email protected] (A.M.d.l.R.-G.); [email protected] (M.P.F.-P.); [email protected] (L.R.-G.); [email protected] (L.Z.-M.); [email protected] (I.R.-L.); [email protected] (V.V.) * Correspondence: [email protected] (R.G.-C.); [email protected] (C.M.); Tel.: +34-968341990 (R.G.-C. & C.M.); Fax: +34-968261914 (R.G.-C. & C.M.) † These authors contributed equally to this work. ‡ These authors shared senior authorship. Abstract: Neutrophil extracellular traps (NETs) are formed after neutrophils expelled their chromatin content in order to primarily capture and eliminate pathogens. However, given their characteristics due in part to DNA and different granular proteins, NETs may induce a procoagulant response linking inflammation and thrombosis. Unraveling NET formation molecular mechanisms as well Citation: Águila, S.; de los as the intracellular elements that regulate them is relevant not only for basic knowledge but also Reyes-García, A.M.; Fernández-Pérez, to design diagnostic and therapeutic tools that may prevent their deleterious effects observed in M.P.; Reguilón-Gallego, L.; several inflammatory pathologies (e.g., cardiovascular and autoimmune diseases, cancer). -

Identification of Key Pathways and Genes in Dementia Via Integrated Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of Key Pathways and Genes in Dementia via Integrated Bioinformatics Analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract To provide a better understanding of dementia at the molecular level, this study aimed to identify the genes and key pathways associated with dementia by using integrated bioinformatics analysis. Based on the expression profiling by high throughput sequencing dataset GSE153960 derived from the Gene Expression Omnibus (GEO), the differentially expressed genes (DEGs) between patients with dementia and healthy controls were identified. With DEGs, we performed a series of functional enrichment analyses. Then, a protein–protein interaction (PPI) network, modules, miRNA-hub gene regulatory network and TF-hub gene regulatory network was constructed, analyzed and visualized, with which the hub genes miRNAs and TFs nodes were screened out. Finally, validation of hub genes was performed by using receiver operating characteristic curve (ROC) analysis. -

Characterization of the Macrophage Transcriptome in Glomerulonephritis-Susceptible and -Resistant Rat Strains
Genes and Immunity (2011) 12, 78–89 & 2011 Macmillan Publishers Limited All rights reserved 1466-4879/11 www.nature.com/gene ORIGINAL ARTICLE Characterization of the macrophage transcriptome in glomerulonephritis-susceptible and -resistant rat strains K Maratou1, J Behmoaras2, C Fewings1, P Srivastava1, Z D’Souza1, J Smith3, L Game4, T Cook2 and T Aitman1 1Physiological Genomics and Medicine Group, MRC Clinical Sciences Centre, Imperial College London, London, UK; 2Centre for Complement and Inflammation Research, Imperial College London, London, UK; 3Renal Section, Imperial College London, London, UK and 4Genomics Laboratory, MRC Clinical Sciences Centre, London, UK Crescentic glomerulonephritis (CRGN) is a major cause of rapidly progressive renal failure for which the underlying genetic basis is unknown. Wistar–Kyoto (WKY) rats show marked susceptibility to CRGN, whereas Lewis rats are resistant. Glomerular injury and crescent formation are macrophage dependent and mainly explained by seven quantitative trait loci (Crgn1–7). Here, we used microarray analysis in basal and lipopolysaccharide (LPS)-stimulated macrophages to identify genes that reside on pathways predisposing WKY rats to CRGN. We detected 97 novel positional candidates for the uncharacterized Crgn3–7. We identified 10 additional secondary effector genes with profound differences in expression between the two strains (45-fold change, o1% false discovery rate) for basal and LPS-stimulated macrophages. Moreover, we identified eight genes with differentially expressed alternatively spliced isoforms, by using an in-depth analysis at the probe level that allowed us to discard false positives owing to polymorphisms between the two rat strains. Pathway analysis identified several common linked pathways, enriched for differentially expressed genes, which affect macrophage activation. -

Editorial: Functions of Non-Coding RNA in Innate Immunity
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Frontiers - Publisher Connector EDITORIAL published: 14 December 2015 doi: 10.3389/fimmu.2015.00622 Editorial: Functions of Non-Coding RNA in Innate Immunity Susan Carpenter* Department of Molecular, Cell and Developmental Biology, University of California, Santa Cruz, Santa Cruz, CA, USA Keywords: innate immunity, long non-coding RNAs, miRNAs, inflammation, extracellular RNA Our understanding of the functions that non-coding RNAs play in shaping the immune response is still in its infancy. The breakthroughs in deep sequencing technology have provided us with an unprecedented view of the human genome. The deeper we sequence the more non-coding genes we identify, while the number of protein coding genes remains constant. GENCODE represents the gene set of the ENCODE project and there are currently 15,931 long non-coding RNA (lncRNA) genes, 9,882 small non-coding RNA genes, and 14,477 Pseudogenes cataloged in GENCODE version 231. The contribution of each of these genes to biological processes still remains to be determined. In this research topic, we explore recent data surrounding the functions for microRNA (miRNA) as well as lncRNA within Innate Immunity. Innate immune responses to infection involve the production of pro-inflammatory cytokines such as IL-6 and TNFα in addition to the Type I Interferons (IFNs) that play critical roles in anti-viral immunity. In recent years, there have been huge strides made in understanding the contributions of small RNAs such as miRNA to the Innate Immune processes. More recently, our attention has also been drawn to the growing catalog of lncRNAs. -

Identification of a Microdeletion at Xp22.13 in a Taiwanese Family
Journal of Human Genetics (2011) 56, 8–11 & 2011 The Japan Society of Human Genetics All rights reserved 1434-5161/11 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Identification of a microdeletion at Xp22.13 in a Taiwanese family presenting with Nance-Horan syndrome Hsiao-Mei Liao1, Dau-Ming Niu2,3, Yan-Jang Chen4,5, Jye-Siung Fang6, Shih-Jen Chen7 and Chia-Hsiang Chen8,9,10 Nance-Horan syndrome (NHS) is a rare X-linked disorder characterized by congenital cataracts, dental anomalies and mental retardation. The disease has been linked to a novel gene termed NHS located at Xp22.13. The majority of pathogenic mutations of the disease include nonsense mutations and small deletions and insertions that lead to truncation of the NHS protein. In this study, we identified a microdeletion of B0.92 Mb at Xp22.13 detected by array-based comparative genomic hybridization in two brothers presenting congenital cataract, dental anomalies, facial dysmorphisms and mental retardation. The deleted region encompasses the REPS2, NHS, SCML1 and RAI2 genes, and was transmitted from their carrier mother who presented only mild cataract. Our findings are in line with several recent case reports to indicate that genomic rearrangement involving the NHS gene is an important genetic etiology underlying NHS. Journal of Human Genetics (2011) 56, 8–11; doi:10.1038/jhg.2010.121; published online 30 September 2010 Keywords: array CGH; congenital cataract; mental retardation; microdeletion; Nance-Horan syndrome INTRODUCTION associated with patient with NHS or X-linked congenital cataract,6,12 Nance-Horan syndrome (NHS) (OMIM 302350) is a rare X-linked suggesting that genomic rearrangement also contributes to the genetic genetic disorder characterized by congenital cataracts, dental defects, etiology underlying the NHS. -

REPS2 Antibody (N-Term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap13131a
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 REPS2 Antibody (N-term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP13131a Specification REPS2 Antibody (N-term) - Product Information Application WB, IHC-P,E Primary Accession Q8NFH8 Other Accession NP_004717.2, NP_001074444.1 Reactivity Human, Mouse Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 71534 Antigen Region 153-182 REPS2 Antibody (N-term) - Additional Information REPS2 Antibody (N-term) (Cat. #AP13131a) Gene ID 9185 western blot analysis in mouse brain tissue lysates (35ug/lane).This demonstrates the Other Names REPS2 antibody detected the REPS2 protein RalBP1-associated Eps domain-containing (arrow). protein 2, Partner of RalBP1, RalBP1-interacting protein 2, REPS2, POB1 Target/Specificity This REPS2 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 153-182 amino acids from the N-terminal region of human REPS2. Dilution WB~~1:1000 IHC-P~~1:10~50 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. REPS2 Antibody (N-term) (Cat. #AP13131a)immunohistochemistry analysis Storage in formalin fixed and paraffin embedded Maintain refrigerated at 2-8°C for up to 2 human cerebellum tissue followed by weeks. For long term storage store at -20°C peroxidase conjugation of the secondary in small aliquots to prevent freeze-thaw antibody and DAB staining.This data cycles. demonstrates the use of REPS2 Antibody (N-term) for immunohistochemistry. Clinical Precautions relevance has not been evaluated. -

Combined Effects of PVT1 and Mir-146A Single Nucleotide
Int. J. Biol. Sci. 2018, Vol. 14 1153 Ivyspring International Publisher International Journal of Biological Sciences 2018; 14(10): 1153-1162. doi: 10.7150/ijbs.25420 Research Paper Combined Effects of PVT1 and MiR-146a Single Nucleotide Polymorphism on the Lung Function of Smokers with Chronic Obstructive Pulmonary Disease Sijing Zhou1*, Yi Liu2*, Min Li3, Peipei Wu2, Gengyun Sun2, Guanghe Fei2, Xuan Xu4, Xuexin Zhou5, Luqian Zhou5, Ran Wang2 1. Hefei Prevention and Treatment Center for Occupational Diseases, Hefei 230022, China 2. Department of respiratory and critical care medicine, the first affiliated hospital of Anhui medical university, Hefei 230022, China 3. Department of oncology, the first affiliated hospital of Anhui medical university, Hefei 230022, China 4. Division of Pulmonary/Critical Care Medicine, Cedars sinai Medical Center, Los Angeles 90015, USA 5. The first clinical college of Anhui medical university, Hefei 230032, China *These authors contributed equally to the work. Corresponding author: Ran Wang Ph.D., Department of respiratory and critical care medicine, The first affiliated hospital of Anhui medical university, Hefei, 230022, China. Phone: 86-551-62922913; Fax: 86-551-62922913; E-mail: [email protected] © Ivyspring International Publisher. This is an open access article distributed under the terms of the Creative Commons Attribution (CC BY-NC) license (https://creativecommons.org/licenses/by-nc/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2018.02.07; Accepted: 2018.06.16; Published: 2018.06.23 Abstract Non-coding RNAs play an important role in the pathogenesis of chronic obstructive pulmonary disease (COPD). This study was performed to investigate the role of PVT1 and miR-146a single nucleotide polymorphisms (SNPs) in the lung function of COPD smokers.