Use of Yeast Chemigenomics and COXEN Informatics in Preclinical
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Binnenwerk Cindy Postma.Indd
CHAPTER 6 Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression Gut 2009, 58: 79-89 Beatriz Carvalho Cindy Postma Sandra Mongera Erik Hopmans Sharon Diskin Mark A. van de Wiel Wim van Criekinge Olivier Thas Anja Matthäi Miguel A. Cuesta Jochim S. Terhaar sive Droste Mike Craanen Evelin Schröck Bauke Ylstra Gerrit A. Meijer 104 | Chapter 6 Abstract Objective: This study aimed to identify the oncogenes at 20q involved in colorectal adenoma to carcinoma progression by measuring the effect of 20q gain on mRNA expression of genes in this amplicon. Methods: Segmentation of DNA copy number changes on 20q was performed by array CGH in 34 non-progressed colorectal adenomas, 41 progressed adenomas (i.e. adenomas that present a focus of cancer) and 33 adenocarcinomas. Moreover, a robust analysis of altered expression of genes in these segments was performed by microarray analysis in 37 adenomas and 31 adenocarcinomas. Protein expression was evaluated by immunohistochemistry on tissue microarrays. Results: The genes C20orf24, AURKA, RNPC1, TH1L, ADRM1, C20orf20 and TCFL5, mapping at 20q were signifi cantly overexpressed in carcinomas compared to adenomas as consequence of copy number gain of 20q. Conclusion: This approach revealed C20orf24, AURKA, RNPC1, TH1L, ADRM1, C20orf20 and TCFL5 genes to be important in chromosomal instability-related adenoma to carcinoma progression. These genes therefore may serve as highly specifi c biomarkers for colorectal cancer with potential clinical applications. Putative oncogenes at chromosome 20q in colorectal carcinogenesis | 105 Introduction The majority of cancers are epithelial in origin and arise through a stepwise progression from normal cells, through dysplasia, into malignant cells that invade surrounding tissues and have metastatic potential. -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

High-Throughput, Pooled Sequencing Identifies Mutations in NUBPL And
ARTICLES High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency Sarah E Calvo1–3,10, Elena J Tucker4,5,10, Alison G Compton4,10, Denise M Kirby4, Gabriel Crawford3, Noel P Burtt3, Manuel Rivas1,3, Candace Guiducci3, Damien L Bruno4, Olga A Goldberger1,2, Michelle C Redman3, Esko Wiltshire6,7, Callum J Wilson8, David Altshuler1,3,9, Stacey B Gabriel3, Mark J Daly1,3, David R Thorburn4,5 & Vamsi K Mootha1–3 Discovering the molecular basis of mitochondrial respiratory chain disease is challenging given the large number of both mitochondrial and nuclear genes that are involved. We report a strategy of focused candidate gene prediction, high-throughput sequencing and experimental validation to uncover the molecular basis of mitochondrial complex I disorders. We created seven pools of DNA from a cohort of 103 cases and 42 healthy controls and then performed deep sequencing of 103 candidate genes to identify 151 rare variants that were predicted to affect protein function. We established genetic diagnoses in 13 of 60 previously unsolved cases using confirmatory experiments, including cDNA complementation to show that mutations in NUBPL and FOXRED1 can cause complex I deficiency. Our study illustrates how large-scale sequencing, coupled with functional prediction and experimental validation, can be used to identify causal mutations in individual cases. Complex I of the mitochondrial respiratory chain is a large ~1-MDa assembly factors are probably required, as suggested by the 20 factors macromolecular machine composed of 45 protein subunits encoded necessary for assembly of the smaller complex IV9 and by cohort by both the nuclear and mitochondrial (mtDNA) genomes. -

Differential Expression Profile Prioritization of Positional Candidate Glaucoma Genes the GLC1C Locus
LABORATORY SCIENCES Differential Expression Profile Prioritization of Positional Candidate Glaucoma Genes The GLC1C Locus Frank W. Rozsa, PhD; Kathleen M. Scott, BS; Hemant Pawar, PhD; John R. Samples, MD; Mary K. Wirtz, PhD; Julia E. Richards, PhD Objectives: To develop and apply a model for priori- est because of moderate expression and changes in tization of candidate glaucoma genes. expression. Transcription factor ZBTB38 emerges as an interesting candidate gene because of the overall expres- Methods: This Affymetrix GeneChip (Affymetrix, Santa sion level, differential expression, and function. Clara, Calif) study of gene expression in primary cul- ture human trabecular meshwork cells uses a positional Conclusions: Only1geneintheGLC1C interval fits our differential expression profile model for prioritization of model for differential expression under multiple glau- candidate genes within the GLC1C genetic inclusion in- coma risk conditions. The use of multiple prioritization terval. models resulted in filtering 7 candidate genes of higher interest out of the 41 known genes in the region. Results: Sixteen genes were expressed under all condi- tions within the GLC1C interval. TMEM22 was the only Clinical Relevance: This study identified a small sub- gene within the interval with differential expression in set of genes that are most likely to harbor mutations that the same direction under both conditions tested. Two cause glaucoma linked to GLC1C. genes, ATP1B3 and COPB2, are of interest in the con- text of a protein-misfolding model for candidate selec- tion. SLC25A36, PCCB, and FNDC6 are of lesser inter- Arch Ophthalmol. 2007;125:117-127 IGH PREVALENCE AND PO- identification of additional GLC1C fami- tential for severe out- lies7,18-20 who provide optimal samples for come combine to make screening candidate genes for muta- adult-onset primary tions.7,18,20 The existence of 2 distinct open-angle glaucoma GLC1C haplotypes suggests that muta- (POAG) a significant public health prob- tions will not be limited to rare descen- H1 lem. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Supplementary Table 2 Gene Sets Used in GSEA
Supplementary Table 2 Gene sets used in GSEA Up in RNAi and Sign Confirmed in Inducible Gene Probe Set ID Accession Symbol Gene Title 200660_at NM_005620 S100A11 S100 calcium binding protein A11 (calgizzarin) 200785_s_at NM_002332 LRP1 low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) 201325_s_at NM_001423 EMP1 epithelial membrane protein 1 201373_at NM_000445 PLEC1 plectin 1, intermediate filament binding protein 500kDa 201466_s_at NM_002228 JUN v-jun sarcoma virus 17 oncogene homolog (avian) 201952_at AA156721 ALCAM activated leukocyte cell adhesion molecule 202042_at NM_002109 HARS histidyl-tRNA synthetase 202074_s_at NM_021980 OPTN optineurin 202087_s_at NM_001912 CTSL cathepsin L 202588_at NM_000476 AK1 adenylate kinase 1 202609_at NM_004447 EPS8 epidermal growth factor receptor pathway substrate 8 202733_at NM_004199 P4HA2 procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide II 202756_s_at NM_002081 GPC1 glypican 1 202786_at NM_013233 STK39 serine threonine kinase 39 (STE20/SPS1 homolog, yeast) 202859_x_at NM_000584 IL8 interleukin 8 203083_at NM_003247 THBS2 thrombospondin 2 203186_s_at NM_002961 S100A4 S100 calcium binding protein A4 (calcium protein, calvasculin, metastasin, murine placental homolog) 203232_s_at NM_000332 ATXN1 ataxin 1 203233_at NM_000418 IL4R interleukin 4 receptor 203771_s_at AA740186 BLVRA biliverdin reductase A 203821_at NM_001945 HBEGF heparin-binding EGF-like growth factor 203939_at NM_002526 NT5E 5'-nucleotidase, ecto (CD73) 203955_at NM_014811 -
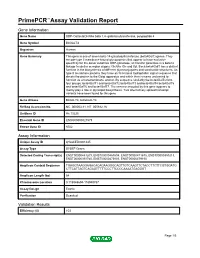
Download Validation Data
PrimePCR™Assay Validation Report Gene Information Gene Name UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 Gene Symbol B4GALT4 Organism Human Gene Summary This gene is one of seven beta-14-galactosyltransferase (beta4GalT) genes. They encode type II membrane-bound glycoproteins that appear to have exclusive specificity for the donor substrate UDP-galactose; all transfer galactose in a beta14 linkage to similar acceptor sugars: GlcNAc Glc and Xyl. Each beta4GalT has a distinct function in the biosynthesis of different glycoconjugates and saccharide structures. As type II membrane proteins they have an N-terminal hydrophobic signal sequence that directs the protein to the Golgi apparatus and which then remains uncleaved to function as a transmembrane anchor. By sequence similarity the beta4GalTs form four groups: beta4GalT1 and beta4GalT2 beta4GalT3 and beta4GalT4 beta4GalT5 and beta4GalT6 and beta4GalT7. The enzyme encoded by this gene appears to mainly play a role in glycolipid biosynthesis. Two alternatively spliced transcript variants have been found for this gene. Gene Aliases B4Gal-T4, beta4Gal-T4 RefSeq Accession No. NC_000003.11, NT_005612.16 UniGene ID Hs.13225 Ensembl Gene ID ENSG00000121578 Entrez Gene ID 8702 Assay Information Unique Assay ID qHsaCED0001445 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000483209, ENST00000467604, ENST00000471675, ENST00000359213, ENST00000393765, ENST00000475803, ENST00000479150 Amplicon Context Sequence TGAGGTAAGGAGACACAGAAGGGCAGTTGTCAAGTTCTACCTTCTTCGTGGATG CTTCATTAGTCAGAGTTTTTCCCTTCCCCAAAATGAGGGT Amplicon Length (bp) 64 Chromosome Location 3:118948694-118948787 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 103 Page 1/5 PrimePCR™Assay Validation Report R2 0.9998 cDNA Cq 21.23 cDNA Tm (Celsius) 78 gDNA Cq 23.9 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. -

A Crosstalk Between the RNA Binding Protein Smaug and the Hedgehog Pathway Links Cell Signaling to Mrna Regulation in Drosophila Lucía Bruzzone
A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila Lucía Bruzzone To cite this version: Lucía Bruzzone. A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila. Cellular Biology. Université Sorbonne Paris Cité, 2018. English. NNT : 2018USPCC234. tel-02899776 HAL Id: tel-02899776 https://tel.archives-ouvertes.fr/tel-02899776 Submitted on 15 Jul 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Thèse de doctorat de l’Université Sorbonne Paris Cité Préparée à l’Université Paris Diderot Ecole doctorale HOB n° 561 Institut Jacques Monod / Equipe Développement, Signalisation et Trafic A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in Drosophila Lucía Bruzzone Thèse de doctorat de Biologie Dirigée par Anne Plessis Présentée et soutenue publiquement à Paris le 19 mars 2018 Président du jury: Alain Zider / Professeur Université Paris Diderot -

(12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer Et Al
USOO8440393B2 (12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer et al. (45) Date of Patent: May 14, 2013 (54) PRO-ANGIOGENIC GENES IN OVARIAN OTHER PUBLICATIONS TUMORENDOTHELIAL CELL, SOLATES Boyd (The Basic Science of Oncology, 1992, McGraw-Hill, Inc., p. (75) Inventors: Michael J. Birrer, Mt. Airy, MD (US); 379). Tomas A. Bonome, Washington, DC Tockman et al. (Cancer Res., 1992, 52:2711s-2718s).* (US); Anil Sood, Pearland, TX (US); Pritzker (Clinical Chemistry, 2002, 48: 1147-1150).* Chunhua Lu, Missouri City, TX (US) Benedict et al. (J. Exp. Medicine, 2001, 193(1) 89-99).* Jiang et al. (J. Biol. Chem., 2003, 278(7) 4763-4769).* (73) Assignees: The United States of America as Matsushita et al. (FEBS Letters, 1999, vol. 443, pp. 348-352).* Represented by the Secretary of the Singh et al. (Glycobiology, 2001, vol. 11, pp. 587-592).* Department of Health and Human Abbosh et al. (Cancer Res. Jun. 1, 2006 66:5582-55.91 and Supple Services, Washington, DC (US); The mental Figs. S1-S7).* University of MD Anderson Cancer Zhai et al. (Chinese General Practice Aug. 2008, 11(8A): 1366 Center, Houston, TX (US) 1367).* Lu et al. (Cancer Res. Feb. 15, 2007, 64(4): 1757-1768).* (*) Notice: Subject to any disclaimer, the term of this Bagnato et al., “Activation of Mitogenic Signaling by Endothelin 1 in patent is extended or adjusted under 35 Ovarian Carcinoma Cells', Cancer Research, vol. 57, pp. 1306-1311, U.S.C. 154(b) by 194 days. 1997. Bouras et al., “Stanniocalcin 2 is an Estrogen-responsive Gene (21) Appl. -

Tese Roberto Molina.Pdf
Universidade de São Paulo FACULDADE DE MEDICINA DE RIBEIRÃO PRETO Roberto Augusto Silva Molina Tese de Doutorado a ser apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para a obtenção do Título de Doutor em Ciências: Biologia Celular e Molecular. Orientadora: Prof.ª Dr.ª Enilza Maria Espreafico RIBEIRÃO PRETO 2014 Ficha Catalográfica Molina, Roberto Augusto Silva "EMC1 contribui para a malignidade em células de câncer de mama", Ribeirão Preto, 2014. 107p.: il.; 30cm Tese de Doutorado, apresentada à Faculdade de Medicina de Ribeirão Preto/USP. Área de Concentração: Biologia Celular e Molecular. Orientadora: Espreafico, Enilza Maria. 1 - EMC1; 2 - KIAA0090; 3 - câncer de mama; 4 - invasão; 5 - EROs FOLHA DE APROVAÇÃO Roberto Augusto Silva Molina "EMC1 contribui para a malignidade em células de câncer de mama" Tese de doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para a obtenção do título de Doutor. Área de Concentração: Biologia Celular e Molecular. Aprovado em: / / Banca Examinadora Professora Dra. Maria Aparecida Nagai Instituição: Assinatura: ________________________________________________________ Professora Dra. Mariana Kiomy Osako Instituição: Assinatura: ________________________________________________________ Professor Dr. Guido Lenz Instituição: Assinatura: ________________________________________________________ Professor Dr. Wilson Araujo da Silva Júnior Instituição: Assinatura: ________________________________________________________ Professora Dra. Enilza Maria Espreafico Instituição: Faculdade de Medicina de Ribeirão Preto/USP Assinatura: ________________________________________________________ i Agradecimentos... Primeiramente a Deus, que me concedeu o dom da vida, por iluminar meu caminho sempre me dando forças para lutar sem perder o ânimo. À Profa. Dra. Enilza Maria Espreafico, por me receber de portas abertas em seu laboratório, pela confiança depositada, por todo ensinamento e pela sua empolgação contagiante pela ciência. -

Supplemental Table 3 Site ID Intron Poly(A) Site Type NM/KG Inum
Supplemental Table 3 Site ID Intron Poly(A) site Type NM/KG Inum Region Gene ID Gene Symbol Gene Annotation Hs.120277.1.10 chr3:170997234:170996860 170996950 b NM_153353 7 CDS 151827 LRRC34 leucine rich repeat containing 34 Hs.134470.1.27 chr17:53059664:53084458 53065543 b NM_138962 10 CDS 124540 MSI2 musashi homolog 2 (Drosophila) Hs.162889.1.18 chr14:80367239:80329208 80366262 b NM_152446 12 CDS 145508 C14orf145 chromosome 14 open reading frame 145 Hs.187898.1.27 chr22:28403623:28415294 28404458 b NM_181832 16 3UTR 4771 NF2 neurofibromin 2 (bilateral acoustic neuroma) Hs.228320.1.6 chr10:115527009:115530350 115527470 b BC036365 5 CDS 79949 C10orf81 chromosome 10 open reading frame 81 Hs.266308.1.2 chr11:117279579:117278191 117278967 b NM_032046 12 CDS 84000 TMPRSS13 transmembrane protease, serine 13 Hs.266308.1.4 chr11:117284536:117281662 117283722 b NM_032046 9 CDS 84000 TMPRSS13 transmembrane protease, serine 13 Hs.2689.1.4 chr10:53492398:53563605 53492622 b NM_006258 7 CDS 5592 PRKG1 protein kinase, cGMP-dependent, type I Hs.280781.1.6 chr18:64715646:64829150 64715837 b NM_024781 4 CDS 79839 C18orf14 chromosome 18 open reading frame 14 Hs.305985.2.25 chr12:8983686:8984438 8983942 b BX640639 17 3UTR NA NA NA Hs.312098.1.36 chr1:151843991:151844258 151844232 b NM_003815 15 CDS 8751 ADAM15 a disintegrin and metalloproteinase domain 15 (metargidin) Hs.314338.1.11 chr21:39490293:39481214 39487623 b NM_018963 41 CDS 54014 BRWD1 bromodomain and WD repeat domain containing 1 Hs.33368.1.3 chr15:92685158:92689361 92688314 b NM_018349 6 CDS 55784 MCTP2 multiple C2-domains with two transmembrane regions 2 Hs.346736.1.21 chr2:99270738:99281614 99272414 b AK126402 10 3UTR 51263 MRPL30 mitochondrial ribosomal protein L30 Hs.445061.1.19 chr16:69322898:69290216 69322712 b NM_018052 14 CDS 55697 VAC14 Vac14 homolog (S. -

Human Genome Center Laboratory of Genome Database Laboratory of Sequence Analysis ゲノムデータベース分野 シークエンスデータ情報処理分野
116 Human Genome Center Laboratory of Genome Database Laboratory of Sequence Analysis ゲノムデータベース分野 シークエンスデータ情報処理分野 Professor Minoru Kanehisa, Ph.D. 教授(委嘱) 理学博士 金 久 實 Assistant Professor Toshiaki Katayama, M.Sc. 助 教 理学修士 片山俊明 Assistant Professor Shuichi Kawashima, M.Sc. 助 教 理学修士 川島秀一 Lecturer Tetsuo Shibuya, Ph.D. 講 師 理学博士 渋谷哲朗 Assistant Professor Michihiro Araki, Ph.D. 助 教 薬学博士 荒木通啓 Since the completion of the Human Genome Project, high-throughput experimental projects have been initiated for uncovering genomic information in an extended sense, including transcriptomics, proteomics metabolomics, glycomics, and chemi- cal genomics. We are developing a new generation of databases and computa- tional technologies, beyond the traditional genome databases and sequence analy- sis tools, for making full use of these divergent and ever-increasing amounts of data, especially for medical and pharmaceutical applications. 1. KEGG DRUG and KEGG DISEASE natural product information in the context of plant and other genomes. We also develop Minoru Kanehisa KEGG DISEASE (http://www.genome.jp/kegg/ disease/), a new resource for understanding KEGG is a database of biological systems that molecular mechanisms of human diseases, integrates genomic, chemical, and systemic func- where molecular networks involving disease tional information. It is widely used as a refer- genes are represented as KEGG pathway maps ence knowledge base for understanding higher- or, when such details are not known, simply by order functions and utilities of the cell or the or- a list of diseases genes and other lists of mole- ganism from genomic information. Although the cules including environmental factors. basic components of the KEGG resource are de- veloped in Kyoto University, this Laboratory in 2.