Genomic Evidence of Geographically Widespread Effect of Gene Flow From
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A
s l a m m a y t T i M S N v I i A e G t A n i p E S r a A C a C E H n T M i THE CASE AGAINST Marine Mammals in Captivity The Humane Society of the United State s/ World Society for the Protection of Animals 2009 1 1 1 2 0 A M , n o t s o g B r o . 1 a 0 s 2 u - e a t i p s u S w , t e e r t S h t u o S 9 8 THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A. Rose, E.C.M. Parsons, and Richard Farinato, 4th edition Editors: Naomi A. Rose and Debra Firmani, 4th edition ©2009 The Humane Society of the United States and the World Society for the Protection of Animals. All rights reserved. ©2008 The HSUS. All rights reserved. Printed on recycled paper, acid free and elemental chlorine free, with soy-based ink. Cover: ©iStockphoto.com/Ying Ying Wong Overview n the debate over marine mammals in captivity, the of the natural environment. The truth is that marine mammals have evolved physically and behaviorally to survive these rigors. public display industry maintains that marine mammal For example, nearly every kind of marine mammal, from sea lion Iexhibits serve a valuable conservation function, people to dolphin, travels large distances daily in a search for food. In learn important information from seeing live animals, and captivity, natural feeding and foraging patterns are completely lost. -
Crystal Reports Activex Designer
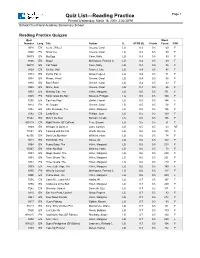
Quiz List—Reading Practice Page 1 Printed Wednesday, March 18, 2009 2:36:33PM School: Churchland Academy Elementary School Reading Practice Quizzes Quiz Word Number Lang. Title Author IL ATOS BL Points Count F/NF 9318 EN Ice Is...Whee! Greene, Carol LG 0.3 0.5 59 F 9340 EN Snow Joe Greene, Carol LG 0.3 0.5 59 F 36573 EN Big Egg Coxe, Molly LG 0.4 0.5 99 F 9306 EN Bugs! McKissack, Patricia C. LG 0.4 0.5 69 F 86010 EN Cat Traps Coxe, Molly LG 0.4 0.5 95 F 9329 EN Oh No, Otis! Frankel, Julie LG 0.4 0.5 97 F 9333 EN Pet for Pat, A Snow, Pegeen LG 0.4 0.5 71 F 9334 EN Please, Wind? Greene, Carol LG 0.4 0.5 55 F 9336 EN Rain! Rain! Greene, Carol LG 0.4 0.5 63 F 9338 EN Shine, Sun! Greene, Carol LG 0.4 0.5 66 F 9353 EN Birthday Car, The Hillert, Margaret LG 0.5 0.5 171 F 9305 EN Bonk! Goes the Ball Stevens, Philippa LG 0.5 0.5 100 F 7255 EN Can You Play? Ziefert, Harriet LG 0.5 0.5 144 F 9314 EN Hi, Clouds Greene, Carol LG 0.5 0.5 58 F 9382 EN Little Runaway, The Hillert, Margaret LG 0.5 0.5 196 F 7282 EN Lucky Bear Phillips, Joan LG 0.5 0.5 150 F 31542 EN Mine's the Best Bonsall, Crosby LG 0.5 0.5 106 F 901618 EN Night Watch (SF Edition) Fear, Sharon LG 0.5 0.5 51 F 9349 EN Whisper Is Quiet, A Lunn, Carolyn LG 0.5 0.5 63 NF 74854 EN Cooking with the Cat Worth, Bonnie LG 0.6 0.5 135 F 42150 EN Don't Cut My Hair! Wilhelm, Hans LG 0.6 0.5 74 F 9018 EN Foot Book, The Seuss, Dr. -

Inferring Ancestry Across Hybrid Genomes Using Low-Coverage Sequence Data
UC Santa Cruz UC Santa Cruz Previously Published Works Title AD-LIBS: inferring ancestry across hybrid genomes using low-coverage sequence data. Permalink https://escholarship.org/uc/item/04j576r0 Journal BMC bioinformatics, 18(1) ISSN 1471-2105 Authors Schaefer, Nathan K Shapiro, Beth Green, Richard E Publication Date 2017-04-04 DOI 10.1186/s12859-017-1613-0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Schaefer et al. BMC Bioinformatics (2017) 18:203 DOI 10.1186/s12859-017-1613-0 SOFTWARE Open Access AD-LIBS: inferring ancestry across hybrid genomes using low-coverage sequence data Nathan K. Schaefer1,3*, Beth Shapiro2,3 and Richard E. Green1,3 Abstract Background: Inferring the ancestry of each region of admixed individuals’ genomesisusefulinstudiesranging from disease gene mapping to speciation genetics. Current methods require high-coverage genotype data and phased reference panels, and are therefore inappropriate for many data sets. We present a software application, AD-LIBS, that uses a hidden Markov model to infer ancestry across hybrid genomes without requiring variant calling or phasing. This approach is useful for non-model organisms and in cases of low-coverage data, such as ancient DNA. Results: We demonstrate the utility of AD-LIBS with synthetic data. We then use AD-LIBS to infer ancestry in two published data sets: European human genomes with Neanderthal ancestry and brown bear genomes with polar bear ancestry. AD-LIBS correctly infers 87–91% of ancestry in simulations and produces ancestry maps that agree with published results and global ancestry estimates in humans. In brown bears, we find more polar bear ancestry than has been published previously, using both AD-LIBS and an existing software application for local ancestry inference, HAPMIX. -

Docent Manual
2018 Docent Manual Suzi Fontaine, Education Curator Montgomery Zoo and Mann Wildlife Learning Museum 7/24/2018 Table of Contents Docent Information ....................................................................................................................................................... 2 Dress Code................................................................................................................................................................. 9 Feeding and Cleaning Procedures ........................................................................................................................... 10 Docent Self-Evaluation ............................................................................................................................................ 16 Mission Statement .................................................................................................................................................. 21 Education Program Evaluation Form ...................................................................................................................... 22 Education Master Plan ............................................................................................................................................ 23 Animal Diets ............................................................................................................................................................ 25 Mammals .................................................................................................................................................................... -

The History of the Tall Ship Regina Maris
Linfield University DigitalCommons@Linfield Linfield Alumni Book Gallery Linfield Alumni Collections 2019 Dreamers before the Mast: The History of the Tall Ship Regina Maris John Kerr Follow this and additional works at: https://digitalcommons.linfield.edu/lca_alumni_books Part of the Cultural History Commons, and the United States History Commons Recommended Citation Kerr, John, "Dreamers before the Mast: The History of the Tall Ship Regina Maris" (2019). Linfield Alumni Book Gallery. 1. https://digitalcommons.linfield.edu/lca_alumni_books/1 This Book is protected by copyright and/or related rights. It is brought to you for free via open access, courtesy of DigitalCommons@Linfield, with permission from the rights-holder(s). Your use of this Book must comply with the Terms of Use for material posted in DigitalCommons@Linfield, or with other stated terms (such as a Creative Commons license) indicated in the record and/or on the work itself. For more information, or if you have questions about permitted uses, please contact [email protected]. Dreamers Before the Mast, The History of the Tall Ship Regina Maris By John Kerr Carol Lew Simons, Contributing Editor Cover photo by Shep Root Third Edition This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc- nd/4.0/. 1 PREFACE AND A TRIBUTE TO REGINA Steven Katona Somehow wood, steel, cable, rope, and scores of other inanimate materials and parts create a living thing when they are fastened together to make a ship. I have often wondered why ships have souls but cars, trucks, and skyscrapers don’t. -

Sing! 1975 – 2014 Song Index
Sing! 1975 – 2014 song index Song Title Composer/s Publication Year/s First line of song 24 Robbers Peter Butler 1993 Not last night but the night before ... 59th St. Bridge Song [Feelin' Groovy], The Paul Simon 1977, 1985 Slow down, you move too fast, you got to make the morning last … A Beautiful Morning Felix Cavaliere & Eddie Brigati 2010 It's a beautiful morning… A Canine Christmas Concerto Traditional/May Kay Beall 2009 On the first day of Christmas my true love gave to me… A Long Straight Line G Porter & T Curtan 2006 Jack put down his lister shears to join the welders and engineers A New Day is Dawning James Masden 2012 The first rays of sun touch the ocean, the golden rays of sun touch the sea. A Wallaby in My Garden Matthew Hindson 2007 There's a wallaby in my garden… A Whole New World (Aladdin's Theme) Words by Tim Rice & music by Alan Menken 2006 I can show you the world. A Wombat on a Surfboard Louise Perdana 2014 I was sitting on the beach one day when I saw a funny figure heading my way. A.E.I.O.U. Brian Fitzgerald, additional words by Lorraine Milne 1990 I can't make my mind up- I don't know what to do. Aba Daba Honeymoon Arthur Fields & Walter Donaldson 2000 "Aba daba ... -" said the chimpie to the monk. ABC Freddie Perren, Alphonso Mizell, Berry Gordy & Deke Richards 2003 You went to school to learn girl, things you never, never knew before. Abiyoyo Traditional Bantu 1994 Abiyoyo .. -

The Image and Cult of the Black Christ in Colonial Mexico and Central America
City University of New York (CUNY) CUNY Academic Works All Dissertations, Theses, and Capstone Projects Dissertations, Theses, and Capstone Projects 10-2014 Disseminating Devotion: The Image and Cult of the Black Christ in Colonial Mexico and Central America Elena FitzPatrick Sifford Graduate Center, City University of New York How does access to this work benefit ou?y Let us know! More information about this work at: https://academicworks.cuny.edu/gc_etds/332 Discover additional works at: https://academicworks.cuny.edu This work is made publicly available by the City University of New York (CUNY). Contact: [email protected] DISSEMINATING DEVOTION: THE IMAGE AND CULT OF THE BLACK CHRIST IN COLONIAL MEXICO AND CENTRAL AMERICA by ELENA FITZPATRICK SIFFORD A dissertation submitted to the Graduate Faculty in Art History in partial fulfillment of the requirements for the degree of Doctor of Philosophy, The City University of New York 2014 ©2014 ELENA FITZPATRICK SIFFORD All Rights Reserved ii This manuscript has been read and accepted for the Graduate Faculty in Art History in satisfaction of the dissertation requirement for the degree of Doctor of Philosophy. Date Dr. Eloise Quiñones Keber Chair of Examining Committee ____________________ ________________________________________ Date Dr. Claire Bishop Acting Executive Officer ____________________ ________________________________________ ___________________________________ Dr. Amanda Wunder ____________________________________ Dr. Timothy Pugh ____________________________________ Dr. Lauren Kilroy-Ewbank Supervisory Committee THE CITY UNIVERSITY OF NEW YORK iii Abstract DISSEMINATING DEVOTION: THE IMAGE AND CULT OF THE BLACK CHRIST IN COLONIAL MEXICO AND CENTRAL AMERICA by Elena FitzPatrick Sifford Adviser: Dr. Eloise Quiñones Keber Following the conquest of Mexico in 1521, Spanish conquerors and friars considered it their duty to bring Christianity to the New World. -

SMALL CETACEANS, BIG PROBLEMS a Global Review of the Impacts of Hunting on Small Whales, Dolphins and Porpoises
SMALL CETACEANS, BIG PROBLEMS A global review of the impacts of hunting on small whales, dolphins and porpoises A report by Sandra Altherr and Nicola Hodgins Edited by Sue Fisher, Kate O’Connell, D.J. Schubert, and Dave Tilford SMALL CETACEANS, BIG PROBLEMS A global review of the impacts of hunting on small whales, dolphins and porpoises A report by Sandra Altherr and Nicola Hodgins Edited by Sue Fisher, Kate O’Connell, D.J. Schubert, and Dave Tilford ACKNOWLEDGEMENTS The authors would like to thank the following people and institutions for providing helpful input, information and photos: Stefan Austermühle and his organisation Mundo Azul, Conservation India, The Dolphin Project, Astrid Fuchs, Dr. Lindsay Porter, Vivian Romano, and Koen Van Waerebeek. Special thanks also to Ava Rinehart and Alexandra Alberg of the Animal Welfare Institute for the design and layout of this report. GLOSSARY AAWP CMS IMARPE NMFS Abidjan Aquatic Wildlife Partnership Convention on the Conservation of Instituto Del Mar Del Peru National Marine Fisheries Service Migratory Species of Wild Animals ACCOBAMS IUU NOAA Agreement on the Conservation COSWIC Illegal, Unreported and Unregulated National Oceanic and Atmospheric of Cetaceans of the Black Sea, Commission on the Status of Fishing Administration Mediterranean Sea and Contiguous Endangered Wildlife in Canada Atlantic Area IUCN SC CPW International Union for Conservation Scientific Committee (International AQUATIC WILD MEAT Collaborative Partnership on of Nature Whaling Commission) The products derived from aquatic Sustainable Wildlife Management mammals and sea turtles that are IWC SMALL CETACEANS used for subsistence food and DFO International Whaling Commission Small and large cetaceans is traditional uses, including shells, Federal Department of Fisheries & neither a biological, nor political bones and organs, as well as bait for Oceans Ministry (Canada) JCNB classification, but has evolved Greenlandic-Canadian Joint fisheries becoming a widely used semantic DWC Commission for Narwhal and Beluga term. -

2018 Annual Report on Conservation and Science
Annual Report on Conservation and Science INTRODUCTION 2 2018 Annual Report on Conservation and Science The Association of Zoos and Aquariums and its member facilities envision a world where, as a result of their work, all people respect, value, and conserve wildlife and wild places. The 2018 Annual Report on Conservation and Science (ARCS) celebrates the conservation efforts, education programs, green (sustainable) business practices, and research projects of AZA-accredited zoos and aquariums and certified related facilities. Content in ARCS reflects submissions made to annual surveys available through AZA’s website. Each survey topic has been carefully defined to maximize consistency of reporting throughout the AZA community. AZA’s Wildlife Conservation Committee and Research and Technology Committee review all of the relevant submissions by members to promote even greater consistency. Field conservation focuses on efforts having a direct impact on animals and habitats in the wild. Education programming includes those with specific goals and delivery methods, defined content, and a clear primary discipline and target audience. Mission-focused research projects involve application of the scientific method, are hypothesis (or question)-driven, involve systematic data collection, and analysis of those data and draw conclusions from the research process. Green business practices focus on the annual documentation and usage of key resources: energy, fuel for transportation, waste, and water – as well as identification of specific green practices being implemented. AZA is grateful to each member that responded to these surveys. The response rate for each 2018 survey was as follows: » field conservation – 92% » education programming – 59% » green business practices – 65% » mission-focused research – 65% While the 2018 ARCS focuses on individual activities undertaken that year, download the 2018 Highlights (available at: aza.org/annual-report-on-conservation-and-science) to learn more about what the AZA community accomplished together. -
An Update from the Us Small Business Administration Hearing
S. HRG. 115–322 THE STATE OF SMALL BUSINESS IN AMERICA: AN UPDATE FROM THE U.S. SMALL BUSINESS ADMINISTRATION HEARING BEFORE THE COMMITTEE ON SMALL BUSINESS AND ENTREPRENEURSHIP UNITED STATES SENATE ONE HUNDRED FIFTEENTH CONGRESS SECOND SESSION MAY 15, 2018 Printed for the Committee on Small Business and Entrepreneurship ( Available via the World Wide Web: http://www.govinfo.gov U.S. GOVERNMENT PUBLISHING OFFICE 30–925 PDF WASHINGTON : 2018 VerDate Sep 11 2014 14:46 Sep 12, 2018 Jkt 030925 PO 00000 Frm 00001 Fmt 5011 Sfmt 5011 C:\DOCS\30925.TXT SHAUN LAP51NQ082 with DISTILLER COMMITTEE ON SMALL BUSINESS AND ENTREPRENEURSHIP ONE HUNDRED FIFTEENTH CONGRESS JAMES E. RISCH, Idaho, Chairman BENJAMIN L. CARDIN, Maryland, Ranking Member MARCO RUBIO, Florida MARIA CANTWELL, Washington RAND PAUL, Kentucky JEANNE SHAHEEN, NEW HAMPSHIRE TIM SCOTT, South Carolina HEIDI HEITKAMP, North Dakota JONI ERNST, Iowa EDWARD J. MARKEY, Massachusetts JAMES M. INHOFE, Oklahoma CORY A. BOOKER, New Jersey TODD YOUNG, Indiana CHRISTOPHER A. COONS, Delaware MICHAEL B. ENZI, Wyoming MAZIE K. HIRONO, Hawaii MIKE ROUNDS, South Dakota TAMMY DUCKWORTH, Illinois JOHN KENNEDY, Louisiana SKIFFINGTON E. HOLDERNESS, Republican Staff Director SEAN MOORE, Democratic Staff Director (II) VerDate Sep 11 2014 14:46 Sep 12, 2018 Jkt 030925 PO 00000 Frm 00002 Fmt 5904 Sfmt 5904 C:\DOCS\30925.TXT SHAUN LAP51NQ082 with DISTILLER CONTENTS OPENING STATEMENTS Page Risch, Hon. James E., Chairman, and a U.S. Senator from Idaho ..................... 1 Cardin, Hon. Benjamin L., Ranking Member, and a U.S. Senator from Mary- land ........................................................................................................................ 2 WITNESS McMahon, Hon. Linda E., Administrator, U.S. Small Business Administra- tion, Washington, DC .......................................................................................... -

Eliminating%Longcrange%And%“Dangerous”%% Wild%Animals
Fall$ Fall$ 08! ! ! August$$ ! ! 2012% ! ! ! ! Eliminating%! LongCrange%and%“Dangerous”%% ! Wild%Animals%from%Entertainment%! ! Demonstrations%! in%New%York%State%% ! ! ! % ! ! ! ! ! Pace%University%Academy%for%Applied%Environmental%Studies%! ! &%the%Committee%to%Ban%Wild%and%Exotic%Animal%Acts! % ! ! ! ! Michelle!D.!Land,!J.D.1! Pace!University!Academy!for!Applied!Environmental!Studies! 901!Bedford!Road!! Pleasantville,!NY!10570! (914)!773L3091! [email protected]!! ! Committee!to!Ban!Wild!and!Exotic!Animal!Acts! Bob!Funck! Neil!Platt! Louise!Simmons! ! ! ! ! ! ! !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!! 1!With!assistance!from!Pace!University!research!assistants,!Caroline!Craig,!Jessica!Goldstein,!Emilie! Guidat,!Meghan!C.!Marshall!and!Cheri!Neal.!!!! ! TABLE!OF!CONTENTS! ! I. SYNOPSIS!(4)! ! II. ISSUES!WITH!“DANGEROUS”!WILD!ANIMALS!IN!ENTERTAINMENT! ! a. Traveling!Circuses!and!Animal!Confinement!(5)! ! b. Training!LongKRange!Wild!Animal!Performers!(7)! ! c. Consequences!of!Physiological!Stressors!in!Wild!CaPtive!Animals!(9)! ! d. Animal!Welfare!Act!and!Regulation!of!Wild!Animals!(13)! ! e. Public!Safety!Risk!(14)! ! f. Lack!of!Educational!Value!(16)! ! III. WORLDWIDE!AND!DOMESTIC!TREND!TOWARD!NONLANIMAL!CIRCUSES! ! a. History!of!the!Circus:!A!Trend!Toward!Human!Performance!Entertainment! (18)! ! b. Today’s!Circuses!(19)! ! c. Zoos!Phasing!Out!Elephant!Exhibits!(20)! ! d. Primate!SPecific!Bans!(21)! ! e. International!Legislation!to!Restrict!Use!of!Wild!Animals!in!Entertainment! (21)! ! f. United!States!Legislation!to!Restrict!Use!of!Wild!Animals!in!Entertainment! (23)! ! IV. PROPOSED!ACTION!AND!IMPACTS! ! a. Proposed!Legislation!(25)! ! b. Animal!Circuses!RePlaced!by!NonKAnimal!Circuses!(27)! ! c. Economic!Impact!of!Eliminating!LongKRange!Wild!Animals!from!Circuses! (27)! ! V. CONCLUSION!(29)! ! ! 2! ! VI. APPENDICES! ! A. !CIRCUS!CITATIONS!AND!INCIDENTS!(PETA)! ! B. !CIRCUS!ANIMAL!INCIDENTS!(PETA)!! ! C. LETTER!FROM!BLAYNE!DOYLE!TO!WESTCHESTER!COUNTY!BOARD!OF! LEGISLATORS!! ! D. -

Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution James A. Cahill1, Richard E. Green2, Tara L. Fulton1, Mathias Stiller1, Flora Jay3, Nikita Ovsyanikov4, Rauf Salamzade2, John St. John2, Ian Stirling5,6, Montgomery Slatkin3, Beth Shapiro1* 1 Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, California, United States of America, 2 Department of Biomolecular Engineering, University of California Santa Cruz, Santa Cruz, California, United States of America, 3 Department of Integrative Biology, University of California Berkeley, Berkeley, California, United States of America, 4 Wrangel Island State Nature Reserve, Pevek, Russia, 5 Wildlife Research Division, Department of Environment, Edmonton, Canada, 6 Department of Biological Sciences, University of Alberta, Edmonton, Canada Abstract Despite extensive genetic analysis, the evolutionary relationship between polar bears (Ursus maritimus) and brown bears (U. arctos) remains unclear. The two most recent comprehensive reports indicate a recent divergence with little subsequent admixture or a much more ancient divergence followed by extensive admixture. At the center of this controversy are the Alaskan ABC Islands brown bears that show evidence of shared ancestry with polar bears. We present an analysis of genome-wide sequence data for seven polar bears, one ABC Islands brown bear, one mainland Alaskan brown bear, and a black bear (U. americanus), plus recently published datasets from other bears. Surprisingly, we find clear evidence for gene flow from polar bears into ABC Islands brown bears but no evidence of gene flow from brown bears into polar bears. Importantly, while polar bears contributed ,1% of the autosomal genome of the ABC Islands brown bear, they contributed 6.5% of the X chromosome.