Molecular Diversification of Salamanders of the Tropical
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Catalogue of the Amphibians of Venezuela: Illustrated and Annotated Species List, Distribution, and Conservation 1,2César L
Mannophryne vulcano, Male carrying tadpoles. El Ávila (Parque Nacional Guairarepano), Distrito Federal. Photo: Jose Vieira. We want to dedicate this work to some outstanding individuals who encouraged us, directly or indirectly, and are no longer with us. They were colleagues and close friends, and their friendship will remain for years to come. César Molina Rodríguez (1960–2015) Erik Arrieta Márquez (1978–2008) Jose Ayarzagüena Sanz (1952–2011) Saúl Gutiérrez Eljuri (1960–2012) Juan Rivero (1923–2014) Luis Scott (1948–2011) Marco Natera Mumaw (1972–2010) Official journal website: Amphibian & Reptile Conservation amphibian-reptile-conservation.org 13(1) [Special Section]: 1–198 (e180). Catalogue of the amphibians of Venezuela: Illustrated and annotated species list, distribution, and conservation 1,2César L. Barrio-Amorós, 3,4Fernando J. M. Rojas-Runjaic, and 5J. Celsa Señaris 1Fundación AndígenA, Apartado Postal 210, Mérida, VENEZUELA 2Current address: Doc Frog Expeditions, Uvita de Osa, COSTA RICA 3Fundación La Salle de Ciencias Naturales, Museo de Historia Natural La Salle, Apartado Postal 1930, Caracas 1010-A, VENEZUELA 4Current address: Pontifícia Universidade Católica do Río Grande do Sul (PUCRS), Laboratório de Sistemática de Vertebrados, Av. Ipiranga 6681, Porto Alegre, RS 90619–900, BRAZIL 5Instituto Venezolano de Investigaciones Científicas, Altos de Pipe, apartado 20632, Caracas 1020, VENEZUELA Abstract.—Presented is an annotated checklist of the amphibians of Venezuela, current as of December 2018. The last comprehensive list (Barrio-Amorós 2009c) included a total of 333 species, while the current catalogue lists 387 species (370 anurans, 10 caecilians, and seven salamanders), including 28 species not yet described or properly identified. Fifty species and four genera are added to the previous list, 25 species are deleted, and 47 experienced nomenclatural changes. -

Nototriton Nelsoni Is a Moss Salamander Endemic to Cloud Forest in Refugio De Vida Silvestre Texíguat, Located in the Departments of Atlántida and Yoro, Honduras
Nototriton nelsoni is a moss salamander endemic to cloud forest in Refugio de Vida Silvestre Texíguat, located in the departments of Atlántida and Yoro, Honduras. This cryptic species long was confused with N. barbouri, a morphologically similar species now considered endemic to the Sierra de Sulaco in the southern part of the department of Yoro. Like many of its congeners, N. nelsoni rarely is observed in the wild, and is known from just five specimens. Pictured here is the holotype of N. nelsoni, collected above La Liberación in Refugio de Vida Silvestre Texíguat at an elevation of 1,420 m. This salamander is one of many herpetofaunal species endemic to the Cordillera Nombre de Dios. ' © Josiah H. Townsend 909 www.mesoamericanherpetology.com www.eaglemountainpublishing.com Amphibians of the Cordillera Nombre de Dios, Honduras: COI barcoding suggests underestimated taxonomic richness in a threatened endemic fauna JOSIAH H. TOWNSEND1 AND LARRY DAVID WILSON2 1Department of Biology, Indiana University of Pennsylvania, Indiana, Pennsylvania 15705–1081, United States. E-mail: [email protected] (Corresponding author) 2Centro Zamorano de Biodiversidad, Escuela Agrícola Panamericana Zamorano, Departamento de Francisco Morazán, Honduras; 16010 SW 207th Avenue, Miami, Florida 33187-1067, United States. E-mail: [email protected] ABSTRACT: The Cordillera Nombre de Dios is a chain of mountains along the northern coast of Honduras that harbors a high degree of herpetofaunal endemism. We present a preliminary barcode reference library of amphibians from the Cordillera Nombre de Dios, based on sampling at 10 sites from 2008 to 2013. We sequenced 187 samples of 21 nominal taxa for the barcoding locus cytochrome oxidase subunit I (COI), and recovered 28 well-differentiated clades. -

Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca
Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca John F. Lamoreux, Meghan W. McKnight, and Rodolfo Cabrera Hernandez Occasional Paper of the IUCN Species Survival Commission No. 53 Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca John F. Lamoreux, Meghan W. McKnight, and Rodolfo Cabrera Hernandez Occasional Paper of the IUCN Species Survival Commission No. 53 The designation of geographical entities in this book, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of IUCN concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The views expressed in this publication do not necessarily reflect those of IUCN or other participating organizations. Published by: IUCN, Gland, Switzerland Copyright: © 2015 International Union for Conservation of Nature and Natural Resources Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior written permission of the copyright holder. Citation: Lamoreux, J. F., McKnight, M. W., and R. Cabrera Hernandez (2015). Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca. Gland, Switzerland: IUCN. xxiv + 320pp. ISBN: 978-2-8317-1717-3 DOI: 10.2305/IUCN.CH.2015.SSC-OP.53.en Cover photographs: Totontepec landscape; new Plectrohyla species, Ixalotriton niger, Concepción Pápalo, Thorius minutissimus, Craugastor pozo (panels, left to right) Back cover photograph: Collecting in Chamula, Chiapas Photo credits: The cover photographs were taken by the authors under grant agreements with the two main project funders: NGS and CEPF. -

The Origins of Chordate Larvae Donald I Williamson* Marine Biology, University of Liverpool, Liverpool L69 7ZB, United Kingdom
lopmen ve ta e l B Williamson, Cell Dev Biol 2012, 1:1 D io & l l o l g DOI: 10.4172/2168-9296.1000101 e y C Cell & Developmental Biology ISSN: 2168-9296 Research Article Open Access The Origins of Chordate Larvae Donald I Williamson* Marine Biology, University of Liverpool, Liverpool L69 7ZB, United Kingdom Abstract The larval transfer hypothesis states that larvae originated as adults in other taxa and their genomes were transferred by hybridization. It contests the view that larvae and corresponding adults evolved from common ancestors. The present paper reviews the life histories of chordates, and it interprets them in terms of the larval transfer hypothesis. It is the first paper to apply the hypothesis to craniates. I claim that the larvae of tunicates were acquired from adult larvaceans, the larvae of lampreys from adult cephalochordates, the larvae of lungfishes from adult craniate tadpoles, and the larvae of ray-finned fishes from other ray-finned fishes in different families. The occurrence of larvae in some fishes and their absence in others is correlated with reproductive behavior. Adult amphibians evolved from adult fishes, but larval amphibians did not evolve from either adult or larval fishes. I submit that [1] early amphibians had no larvae and that several families of urodeles and one subfamily of anurans have retained direct development, [2] the tadpole larvae of anurans and urodeles were acquired separately from different Mesozoic adult tadpoles, and [3] the post-tadpole larvae of salamanders were acquired from adults of other urodeles. Reptiles, birds and mammals probably evolved from amphibians that never acquired larvae. -

Multi-National Conservation of Alligator Lizards
MULTI-NATIONAL CONSERVATION OF ALLIGATOR LIZARDS: APPLIED SOCIOECOLOGICAL LESSONS FROM A FLAGSHIP GROUP by ADAM G. CLAUSE (Under the Direction of John Maerz) ABSTRACT The Anthropocene is defined by unprecedented human influence on the biosphere. Integrative conservation recognizes this inextricable coupling of human and natural systems, and mobilizes multiple epistemologies to seek equitable, enduring solutions to complex socioecological issues. Although a central motivation of global conservation practice is to protect at-risk species, such organisms may be the subject of competing social perspectives that can impede robust interventions. Furthermore, imperiled species are often chronically understudied, which prevents the immediate application of data-driven quantitative modeling approaches in conservation decision making. Instead, real-world management goals are regularly prioritized on the basis of expert opinion. Here, I explore how an organismal natural history perspective, when grounded in a critique of established human judgements, can help resolve socioecological conflicts and contextualize perceived threats related to threatened species conservation and policy development. To achieve this, I leverage a multi-national system anchored by a diverse, enigmatic, and often endangered New World clade: alligator lizards. Using a threat analysis and status assessment, I show that one recent petition to list a California alligator lizard, Elgaria panamintina, under the US Endangered Species Act often contradicts the best available science. -
Comparative Osteology and Evolution of the Lungless Salamanders, Family Plethodontidae David B
COMPARATIVE OSTEOLOGY AND EVOLUTION OF THE LUNGLESS SALAMANDERS, FAMILY PLETHODONTIDAE DAVID B. WAKE1 ABSTRACT: Lungless salamanders of the family Plethodontidae comprise the largest and most diverse group of tailed amphibians. An evolutionary morphological approach has been employed to elucidate evolutionary rela tionships, patterns and trends within the family. Comparative osteology has been emphasized and skeletons of all twenty-three genera and three-fourths of the one hundred eighty-three species have been studied. A detailed osteological analysis includes consideration of the evolution of each element as well as the functional unit of which it is a part. Functional and developmental aspects are stressed. A new classification is suggested, based on osteological and other char acters. The subfamily Desmognathinae includes the genera Desmognathus, Leurognathus, and Phaeognathus. Members of the subfamily Plethodontinae are placed in three tribes. The tribe Hemidactyliini includes the genera Gyri nophilus, Pseudotriton, Stereochilus, Eurycea, Typhlomolge, and Hemidac tylium. The genera Plethodon, Aneides, and Ensatina comprise the tribe Pleth odontini. The highly diversified tribe Bolitoglossini includes three super genera. The supergenera Hydromantes and Batrachoseps include the nominal genera only. The supergenus Bolitoglossa includes Bolitoglossa, Oedipina, Pseudoeurycea, Chiropterotriton, Parvimolge, Lineatriton, and Thorius. Manculus is considered to be congeneric with Eurycea, and Magnadig ita is congeneric with Bolitoglossa. Two species are assigned to Typhlomolge, which is recognized as a genus distinct from Eurycea. No. new information is available concerning Haptoglossa. Recognition of a family Desmognathidae is rejected. All genera are defined and suprageneric groupings are defined and char acterized. Range maps are presented for all genera. Relationships of all genera are discussed. -

Rediscovery at the Type Locality and Detection of a New Population 1José L
Offcial journal website: Amphibian & Reptile Conservation amphibian-reptile-conservation.org 13(2) [General Section]: 126–132 (e193). Thorius narismagnus (Amphibia: Plethodontidae): rediscovery at the type locality and detection of a new population 1José L. Aguilar-López, 2,*Paulina García-Bañuelos, 3Eduardo Pineda, and 4Sean M. Rovito 1,2,3Red de Biología y Conservación de Vertebrados, Instituto de Ecología, A.C., Carretera antigua a Coatepec 351, El Haya, Xalapa, Veracruz, MEXICO 4Unidad de Genómica Avanzada (Langebio), Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, km 9.6 Libramiento Norte Carretera Irapuato-León, Irapuato, Guanajuato CP 36824, MEXICO Abstract.—Of the 42 Critically Endangered species of plethodontid salamanders that occur in Mexico, thirteen have not been reported in more than ten years. Given the lack of reports since 1976, the minute plethodontid salamander Thorius narismagnus is widely considered as missing. However, this report describes the rediscovery of this minute salamander at the type locality (Volcán San Martín), as well as a new locality on Volcán Santa Marta, 28 km southeast of its previously known distribution, both in the Los Tuxtlas region of Veracruz, Mexico. The localities where T. narismagnus has been found are mature forests in a community reserve on Volcán San Martín and a private reserve on Volcán Santa Marta. The presence of maxillary teeth, generally absent in Thorius, are reported here in some T. narismagnus females. Two efforts which may contribute to the conservation of Thorius narismagnus are the preservation of the cloud forests where this species persists, as well as the determination of the presence and possible effect of chytrid fungus in these populations. -

Pseudoeurycea Naucampatepetl. the Cofre De Perote Salamander Is Endemic to the Sierra Madre Oriental of Eastern Mexico. This
Pseudoeurycea naucampatepetl. The Cofre de Perote salamander is endemic to the Sierra Madre Oriental of eastern Mexico. This relatively large salamander (reported to attain a total length of 150 mm) is recorded only from, “a narrow ridge extending east from Cofre de Perote and terminating [on] a small peak (Cerro Volcancillo) at the type locality,” in central Veracruz, at elevations from 2,500 to 3,000 m (Amphibian Species of the World website). Pseudoeurycea naucampatepetl has been assigned to the P. bellii complex of the P. bellii group (Raffaëlli 2007) and is considered most closely related to P. gigantea, a species endemic to the La specimens and has not been seen for 20 years, despite thorough surveys in 2003 and 2004 (EDGE; www.edgeofexistence.org), and thus it might be extinct. The habitat at the type locality (pine-oak forest with abundant bunch grass) lies within Lower Montane Wet Forest (Wilson and Johnson 2010; IUCN Red List website [accessed 21 April 2013]). The known specimens were “found beneath the surface of roadside banks” (www.edgeofexistence.org) along the road to Las Lajas Microwave Station, 15 kilometers (by road) south of Highway 140 from Las Vigas, Veracruz (Amphibian Species of the World website). This species is terrestrial and presumed to reproduce by direct development. Pseudoeurycea naucampatepetl is placed as number 89 in the top 100 Evolutionarily Distinct and Globally Endangered amphib- ians (EDGE; www.edgeofexistence.org). We calculated this animal’s EVS as 17, which is in the middle of the high vulnerability category (see text for explanation), and its IUCN status has been assessed as Critically Endangered. -
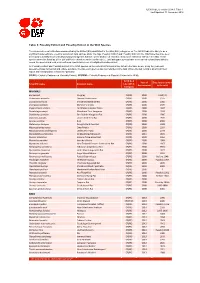
Table 9: Possibly Extinct and Possibly Extinct in the Wild Species
IUCN Red List version 2014.3: Table 9 Last Updated: 13 November 2014 Table 9: Possibly Extinct and Possibly Extinct in the Wild Species The number of recent extinctions documented by the Extinct (EX) and Extinct in the Wild (EW) categories on The IUCN Red List is likely to be a significant underestimate, even for well-known taxa such as birds. The tags 'Possibly Extinct' and 'Possibly Extinct in the Wild' have therefore been developed to identify those Critically Endangered species that are, on the balance of evidence, likely to be extinct (or extinct in the wild). These species cannot be listed as EX or EW until their extinction can be confirmed (i.e., until adequate surveys have been carried out and have failed to record the species and local or unconfirmed reports have been investigated and discounted). All 'Possibly Extinct' and 'Possibly Extinct in the Wild' species on the current IUCN Red List are listed in the table below, along the year each assessment was carried out and, where available, the date each species was last recorded in the wild. Where the last record is an unconfirmed report, last recorded date is noted as "possibly". CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild), IUCN Red Year of Date last recorded Scientific name Common name List (2014) Assessment in the wild Category MAMMALS Bos sauveli Kouprey CR(PE) 2008 1969/70 Crateromys australis Dinagat Crateromys CR(PE) 2008 1975 Crocidura trichura Christmas Island Shrew CR(PE) 2008 1985 Crocidura wimmeri -

Assessing Environmental Variables Across Plethodontid Salamanders
Lauren Mellenthin1, Erica Baken1, Dr. Dean Adams1 1Iowa State University, Ames, Iowa Stereochilus marginatus Pseudotriton ruber Pseudotriton montanus Gyrinophilus subterraneus Gyrinophilus porphyriticus Gyrinophilus palleucus Gyrinophilus gulolineatus Urspelerpes brucei Eurycea tynerensis Eurycea spelaea Eurycea multiplicata Introduction Eurycea waterlooensis Eurycea rathbuni Materials & Methods Eurycea sosorum Eurycea tridentifera Eurycea pterophila Eurycea neotenes Eurycea nana Eurycea troglodytes Eurycea latitans Eurycea naufragia • Arboreality has evolved at least 5 times within Plethodontid salamanders. [1] Eurycea tonkawae Eurycea chisholmensis Climate Variables & Eurycea quadridigitata Polygons & Point Data Eurycea wallacei Eurycea lucifuga Eurycea longicauda Eurycea guttolineata MAXENT Modeling Eurycea bislineata Eurycea wilderae Eurycea cirrigera • Yet no morphological differences separate arboreal and terrestrial species. [1] Eurycea junaluska Hemidactylium scutatum Batrachoseps robustus Batrachoseps wrighti Batrachoseps campi Batrachoseps attenuatus Batrachoseps pacificus Batrachoseps major Batrachoseps luciae Batrachoseps minor Batrachoseps incognitus • There is minimal range overlap between the two microhabitat types. Batrachoseps gavilanensis Batrachoseps gabrieli Batrachoseps stebbinsi Batrachoseps relictus Batrachoseps simatus Batrachoseps nigriventris Batrachoseps gregarius Preliminary results discovered that 71% of the arboreal species distribution Batrachoseps diabolicus Batrachoseps regius Batrachoseps kawia Dendrotriton -

Results of the Global Amphibian Assessment for Bolivia
Results of the Global Amphibian Assessment for Bolivia Diversity Number of Rank in Latin World Rank2 Percentage of Species in Bolivia America and the the World’s Caribbean1 Diversity All Amphibians 201 7 15 3.5 % Frogs & Toads 197 7 14 3.9 % Salamanders 1 13 68 0.2 % Caecilians 3 11 20 1.8 % 1 Out of 44 countries and territories. 2 Out of 192 countries and territories. Threatened Species (Threatened species are in one of the categories in italics.) IUCN Categories Number of Species in IUCN Categories Number of Species in Bolivia Bolivia Extinct 0 Near Threatened 6 Critically Endangered 5 Least Concern 161 Endangered 6 Data Deficient 13 Vulnerable 10 Number and percent of Bolivian species that are threatened: 21 (10%) Extinct Species: none Species that are Critically Endangered and possibly extinct (species that are missing but for which sufficient information is lacking to declare them as extinct): Frogs: Gastrotheca lauzuricae, Hyla chlorostea, Eleutherodactylus zongoensis Role of Protected Areas: Of the 21 threatened species in Bolivia, 76% occur in at least one protected area. Local Experts: Steffen Reichle, The Nature Conservancy, TEL (591-3) 348 0766, 348 0767 Claudia Cortez, Colección Boliviana de Fauna, TEL (591-3) 272 1152 For More Information: Dr. Bruce Young, NatureServe, Costa Rica, TEL (506) 645-6231 Global Amphibian Assessment website: www.globalamphibians.org A detailed analysis of the results for the Americas: www.natureserve.org Results of the Global Amphibian Assessment for Bolivia Map of Amphibian Diversity Source: Global Amphibian Assessment Map of the Distribution of Threatened Species Source: Global Amphibian Assessment Results of the Global Amphibian Assessment for Brazil Diversity Number of Rank in Latin World Rank2 Percentage of Species in Brazil America and the the World’s Caribbean1 Diversity All Amphibians 731 1 1 12.7 % Frogs & Toads 704 1 1 13.9 % Salamanders 1 13 68 0.2 % Caecilians 26 2 2 15.5 % 1 Out of 44 countries and territories. -

Molecular Diversification of Salamanders of The
Blackwell Science, LtdOxford, UKBIJBiological Journal of the Linnean Society0024-4066The Linnean Society of London, 2004? 2004 813 325346 Original Article TROPICAL AMERICAN SALAMANDER DIVERSIFICATION G. PARRA-OLEA ET AL Biological Journal of the Linnean Society, 2004, 81, 325–346. With 7 figures Molecular diversification of salamanders of the tropical American genus Bolitoglossa (Caudata: Plethodontidae) and its evolutionary and biogeographical implications GABRIELA PARRA-OLEA1, MARIO GARCÍA-PARÍS2 and DAVID B. WAKE3* 1Instituto de Biología, UNAM. AP 70–153, CP 04510, Ciudad Universitaria, México D.F., México 2Museo Nacional de Ciencias Naturales, CSIC, José Gutiérrez Abascal, 2, 28006 Madrid, Spain 3Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720–3160, USA Received December 2002; accepted for publication August 2003 The largest genus of salamanders, Bolitoglossa (Plethodontidae), is widespread in tropical America, where it occurs in diverse habitats and elevations, from high elevation grasslands to lowland rain forest. It has the most extensive geographical range of any salamander genus. While most species occur in Middle America, it ranges throughout most of tropical South America as well. Phylogenetic analysis of 1196 bp of two mitochondrial genes (cytochrome b, 16S RNA) from 55 species offers strong support for the monophyly of the genus and sorts the species into a number of clades. Taking into account morphology, distribution, general ecology, and prior systematic and taxonomic studies, we recognize seven subgenera, four of them new: Bolitoglossa Duméril, Bibron et Duméril, 1854, Eladinea Miranda Ribeiro, 1937, Magnadigita Taylor, 1944, Mayamandra, Nanotriton, Oaxakia and Pachymandra. All South American and some lower Middle American species are included in a single well -supported clade, Eladinea.