WES Gene Package Intellectual Disability.Xlsx
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Centronuclear Myopathies Under Attack: a Plethora of Therapeutic Targets Hichem Tasfaout, Belinda Cowling, Jocelyn Laporte
CORE Metadata, citation and similar papers at core.ac.uk Provided by Archive Ouverte en Sciences de l'Information et de la Communication Centronuclear myopathies under attack: A plethora of therapeutic targets Hichem Tasfaout, Belinda Cowling, Jocelyn Laporte To cite this version: Hichem Tasfaout, Belinda Cowling, Jocelyn Laporte. Centronuclear myopathies under attack: A plethora of therapeutic targets. Journal of Neuromuscular Diseases, IOS Press, 2018, 5, pp.387 - 406. 10.3233/JND-180309. hal-02438924 HAL Id: hal-02438924 https://hal.archives-ouvertes.fr/hal-02438924 Submitted on 14 Jan 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Journal of Neuromuscular Diseases 5 (2018) 387–406 387 DOI 10.3233/JND-180309 IOS Press Review Centronuclear myopathies under attack: A plethora of therapeutic targets Hichem Tasfaouta,b,c,d, Belinda S. Cowlinga,b,c,d,1 and Jocelyn Laportea,b,c,d,1,∗ aDepartment of Translational Medicine and Neurogenetics, Institut de G´en´etique et de Biologie Mol´eculaire et Cellulaire (IGBMC), Illkirch, France bInstitut National de la Sant´eetdelaRechercheM´edicale (INSERM), U1258, Illkirch, France cCentre National de la Recherche Scientifique (CNRS), UMR7104, Illkirch, France dUniversit´e de Strasbourg, Illkirch, France Abstract. -

Blueprint Genetics Hereditary Leukemia Panel
Hereditary Leukemia Panel Test code: ON0101 Is a 41 gene panel that includes assessment of non-coding variants. Is ideal for patients with a personal history of a syndrome that confers an increased risk of leukemia or patients with a family history of a syndrome that confers an increased risk of leukemia. About Hereditary Leukemia An inherited predisposition to hematological malignancies, namely acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and bone marrow myelodysplastic syndrome (MDS) may be associated with syndromic features or occur as the principal clinical feature. MDSs and AMLs can occur in the context of syndromic bone marrow failure (eg. dyskeratosis congenita, Fanconi anemia). Other hereditary syndromes with an increased risk of leukemia include Li-Fraumeni syndrome (TP53), ataxia telangiectasia (ATM), Bloom syndrome (BLM), neurofibromatosis type 1 (NF1) and less frequently Noonan syndrome (PTPN11, CBL). Some reports have also shown an association of biallelic germline mutations in constitutional mismatch repair-deficiency syndrome genes, MLH1, MSH2, MSH6, and PMS2 with the development of ALL. Isolated hematological malignancies are associated with germline mutations in RUNX1 (familial platelet syndrome with predisposition to acute myelogenous leukemia), CEBPA (familial AML), GATA2 (GATA2-associated syndromes) and DDX41(DDX41 -related myeloid neoplasms). There is a rapidly expanding list of germline mutations associated with increased risks for myeloid malignancies and inherited predisposition to hematologic malignancies may be more common than has been thought. Many different genetic defects associated with the development of leukemia have been described but the common underlying mechanism is a dysfunctional DNA damage response. Recognition of an inherited cause provides a specific molecular diagnosis and helps to guide treatment, understand unique disease features, prognosis and other organ systems that may be involved, and identify others in the family who may be at risk. -

Affected Female Carriers of MTM1 Mutations Display a Wide Spectrum
Acta Neuropathol DOI 10.1007/s00401-017-1748-0 ORIGINAL PAPER Afected female carriers of MTM1 mutations display a wide spectrum of clinical and pathological involvement: delineating diagnostic clues Valérie Biancalana1,2,3,4,5 · Sophie Scheidecker1 · Marguerite Miguet1 · Annie Laquerrière6 · Norma B. Romero7,8 · Tanya Stojkovic8 · Osorio Abath Neto9 · Sandra Mercier10,11,12 · Nicol Voermans13 · Laura Tanner14 · Curtis Rogers15 · Elisabeth Ollagnon‑Roman16 · Helen Roper17 · Célia Boutte18 · Shay Ben‑Shachar19 · Xavière Lornage2,3,4,5 · Nasim Vasli2,3,4,5 · Elise Schaefer20 · Pascal Laforet21 · Jean Pouget22 · Alexandre Moerman23 · Laurent Pasquier24 · Pascale Marcorelle25,26 · Armelle Magot12 · Benno Küsters27 · Nathalie Streichenberger28 · Christine Tranchant29 · Nicolas Dondaine1 · Raphael Schneider2,3,4,5,30 · Claire Gasnier1 · Nadège Calmels1 · Valérie Kremer31 · Karine Nguyen32 · Julie Perrier12 · Erik Jan Kamsteeg33 · Pierre Carlier34 · Robert‑Yves Carlier35 · Julie Thompson30 · Anne Boland36 · Jean‑François Deleuze36 · Michel Fardeau7,8 · Edmar Zanoteli9 · Bruno Eymard21 · Jocelyn Laporte2,3,4,5 Received: 9 May 2017 / Revised: 24 June 2017 / Accepted: 2 July 2017 © Springer-Verlag GmbH Germany 2017 Abstract X-linked myotubular myopathy (XLMTM), a females and to delineate diagnostic clues, we character- severe congenital myopathy, is caused by mutations in the ized 17 new unrelated afected females and performed a MTM1 gene located on the X chromosome. A majority of detailed comparison with previously reported cases at the afected males die in the early postnatal period, whereas clinical, muscle imaging, histological, ultrastructural and female carriers are believed to be usually asymptomatic. molecular levels. Taken together, the analysis of this large Nevertheless, several afected females have been reported. cohort of 43 cases highlights a wide spectrum of clini- To assess the phenotypic and pathological spectra of carrier cal severity ranging from severe neonatal and generalized weakness, similar to XLMTM male, to milder adult forms. -
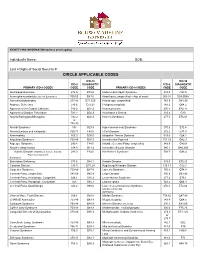
Circle Applicable Codes
IDENTIFYING INFORMATION (please print legibly) Individual’s Name: DOB: Last 4 Digits of Social Security #: CIRCLE APPLICABLE CODES ICD-10 ICD-10 ICD-9 DIAGNOSTIC ICD-9 DIAGNOSTIC PRIMARY ICD-9 CODES CODE CODE PRIMARY ICD-9 CODES CODE CODE Abetalipoproteinemia 272.5 E78.6 Hallervorden-Spatz Syndrome 333.0 G23.0 Acrocephalosyndactyly (Apert’s Syndrome) 755.55 Q87.0 Head Injury, unspecified – Age of onset: 959.01 S09.90XA Adrenaleukodystrophy 277.86 E71.529 Hemiplegia, unspecified 342.9 G81.90 Arginase Deficiency 270.6 E72.21 Holoprosencephaly 742.2 Q04.2 Agenesis of the Corpus Callosum 742.2 Q04.3 Homocystinuria 270.4 E72.11 Agenesis of Septum Pellucidum 742.2 Q04.3 Huntington’s Chorea 333.4 G10 Argyria/Pachygyria/Microgyria 742.2 Q04.3 Hurler’s Syndrome 277.5 E76.01 or 758.33 Aicardi Syndrome 333 G23.8 Hyperammonemia Syndrome 270.6 E72.4 Alcohol Embryo and Fetopathy 760.71 F84.5 I-Cell Disease 272.2 E77.0 Anencephaly 655.0 Q00.0 Idiopathic Torsion Dystonia 333.6 G24.1 Angelman Syndrome 759.89 Q93.5 Incontinentia Pigmenti 757.33 Q82.3 Asperger Syndrome 299.8 F84.5 Infantile Cerebral Palsy, unspecified 343.9 G80.9 Ataxia-Telangiectasia 334.8 G11.3 Intractable Seizure Disorder 345.1 G40.309 Autistic Disorder (Childhood Autism, Infantile 299.0 F84.0 Klinefelter’s Syndrome 758.7 Q98.4 Psychosis, Kanner’s Syndrome) Biotinidase Deficiency 277.6 D84.1 Krabbe Disease 333.0 E75.23 Canavan Disease 330.0 E75.29 Kugelberg-Welander Disease 335.11 G12.1 Carpenter Syndrome 759.89 Q87.0 Larsen’s Syndrome 755.8 Q74.8 Cerebral Palsy, unspecified 343.69 G80.9 -

Genes in Eyecare Geneseyedoc 3 W.M
Genes in Eyecare geneseyedoc 3 W.M. Lyle and T.D. Williams 15 Mar 04 This information has been gathered from several sources; however, the principal source is V. A. McKusick’s Mendelian Inheritance in Man on CD-ROM. Baltimore, Johns Hopkins University Press, 1998. Other sources include McKusick’s, Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders. Baltimore. Johns Hopkins University Press 1998 (12th edition). http://www.ncbi.nlm.nih.gov/Omim See also S.P.Daiger, L.S. Sullivan, and B.J.F. Rossiter Ret Net http://www.sph.uth.tmc.edu/Retnet disease.htm/. Also E.I. Traboulsi’s, Genetic Diseases of the Eye, New York, Oxford University Press, 1998. And Genetics in Primary Eyecare and Clinical Medicine by M.R. Seashore and R.S.Wappner, Appleton and Lange 1996. M. Ridley’s book Genome published in 2000 by Perennial provides additional information. Ridley estimates that we have 60,000 to 80,000 genes. See also R.M. Henig’s book The Monk in the Garden: The Lost and Found Genius of Gregor Mendel, published by Houghton Mifflin in 2001 which tells about the Father of Genetics. The 3rd edition of F. H. Roy’s book Ocular Syndromes and Systemic Diseases published by Lippincott Williams & Wilkins in 2002 facilitates differential diagnosis. Additional information is provided in D. Pavan-Langston’s Manual of Ocular Diagnosis and Therapy (5th edition) published by Lippincott Williams & Wilkins in 2002. M.A. Foote wrote Basic Human Genetics for Medical Writers in the AMWA Journal 2002;17:7-17. A compilation such as this might suggest that one gene = one disease. -

Cardiomyopathy Precision Panel Overview Indications
Cardiomyopathy Precision Panel Overview Cardiomyopathies are a group of conditions with a strong genetic background that structurally hinder the heart to pump out blood to the rest of the body due to weakness in the heart muscles. These diseases affect individuals of all ages and can lead to heart failure and sudden cardiac death. If there is a family history of cardiomyopathy it is strongly recommended to undergo genetic testing to be aware of the family risk, personal risk, and treatment options. Most types of cardiomyopathies are inherited in a dominant manner, which means that one altered copy of the gene is enough for the disease to present in an individual. The symptoms of cardiomyopathy are variable, and these diseases can present in different ways. There are 5 types of cardiomyopathies, the most common being hypertrophic cardiomyopathy: 1. Hypertrophic cardiomyopathy (HCM) 2. Dilated cardiomyopathy (DCM) 3. Restrictive cardiomyopathy (RCM) 4. Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) 5. Isolated Left Ventricular Non-Compaction Cardiomyopathy (LVNC). The Igenomix Cardiomyopathy Precision Panel serves as a diagnostic and tool ultimately leading to a better management and prognosis of the disease. It provides a comprehensive analysis of the genes involved in this disease using next-generation sequencing (NGS) to fully understand the spectrum of relevant genes. Indications The Igenomix Cardiomyopathy Precision Panel is indicated in those cases where there is a clinical suspicion of cardiomyopathy with or without the following manifestations: - Shortness of breath - Fatigue - Arrythmia (abnormal heart rhythm) - Family history of arrhythmia - Abnormal scans - Ventricular tachycardia - Ventricular fibrillation - Chest Pain - Dizziness - Sudden cardiac death in the family 1 Clinical Utility The clinical utility of this panel is: - The genetic and molecular diagnosis for an accurate clinical diagnosis of a patient with personal or family history of cardiomyopathy, channelopathy or sudden cardiac death. -

Ophthalmology
Ophthalmology Information for health professionals MEDICAL GENETIC TESTING FOR OPHTHALMOLOGY Recent technologies, in particularly Next Generation Sequencing (NGS), allows fast, accurate and valuable diagnostic tests. For Ophthalmology, CGC Genetics has an extensive list of medical genetic tests with clinical integration of results by our Medical Geneticists. 1. EXOME SEQUENCING: Exome Sequencing is a very efficient strategy to study most exons of a patient’s genome, unraveling mutations associated with specific disorders or phenotypes. With this diagnostic strategy, patients can be studied with a significantly reduced turnaround time and cost. CGC Genetics has available 2 options for Exome Sequencing: • Whole Exome Sequencing (WES), which analyzes the entire exome (about 20 000 genes); • Disease Exome by CGC Genetics, which analyzes about 6 000 clinically-relevant genes. Any of these can be performed in the index case or in a Trio. 2. NGS PANELS For NGS panels, several genes associated with the same phenotype are simultaneously sequenced. These panels provide increased diagnostic capability with a significantly reduced turnaround time and cost. CGC Genetics has several NGS panels for Ophthalmology that are constantly updated (www.cgcgenetics.com). Any gene studied in exome or NGS panel can also be individually sequenced and analyzed for deletion/duplication events. 3. EXPERTISE IN MEDICAL GENETICS CGC Genetics has Medical Geneticists specialized in genetic counseling for ophthalmological diseases who may advice in choosing the most appropriate -

American Board of Psychiatry and Neurology, Inc
AMERICAN BOARD OF PSYCHIATRY AND NEUROLOGY, INC. CERTIFICATION EXAMINATION IN NEUROLOGY 2015 Content Blueprint (January 13, 2015) Part A Basic neuroscience Number of questions: 120 01. Neuroanatomy 3-5% 02. Neuropathology 3-5% 03. Neurochemistry 2-4% 04. Neurophysiology 5-7% 05. Neuroimmunology/neuroinfectious disease 2-4% 06. Neurogenetics/molecular neurology, neuroepidemiology 2-4% 07. Neuroendocrinology 1-2% 08. Neuropharmacology 4-6% Part B Behavioral neurology, cognition, and psychiatry Number of questions: 80 01. Development through the life cycle 3-5% 02. Psychiatric and psychological principles 1-3% 03. Diagnostic procedures 1-3% 04. Clinical and therapeutic aspects of psychiatric disorders 5-7% 05. Clinical and therapeutic aspects of behavioral neurology 5-7% Part C Clinical neurology (adult and child) The clinical neurology section of the Neurology Certification Examination is comprised of 60% adult neurology questions and 40% child neurology questions. Number of questions: 200 01. Headache disorders 1-3% 02. Pain disorders 1-3% 03. Epilepsy and episodic disorders 1-3% 04. Sleep disorders 1-3% 05. Genetic disorders 1-3% 2015 ABPN Content Specifications Page 1 of 22 Posted: Certification in Neurology AMERICAN BOARD OF PSYCHIATRY AND NEUROLOGY, INC. 06. Congenital disorders 1-3% 07. Cerebrovascular disease 1-3% 08. Neuromuscular diseases 2-4% 09. Cranial nerve palsies 1-3% 10. Spinal cord diseases 1-3% 11. Movement disorders 1-3% 12. Demyelinating diseases 1-3% 13. Neuroinfectious diseases 1-3% 14. Critical care 1-3% 15. Trauma 1-3% 16. Neuro-ophthalmology 1-3% 17. Neuro-otology 1-3% 18. Neurologic complications of systemic diseases 2-4% 19. -

Dyskeratosis Congenita Precision Panel Overview Indications Clinical
Dyskeratosis Congenita Precision Panel Overview Dyskeratosis Congenita (DKC) is a rare, progressive bone marrow failure syndrome characterized by reticulated skin hyperpigmentation, nail dystrophy and oral leukoplakia. Patients usually present with symptoms of skin hyperpigmentation and nail changes during the first decade of life. It is caused by germline mutations in genes regulating telomere maintenance, resulting in very short telomeres. DKC is a genetically heterogeneous with X-linked recessive form being the most common, autosomal dominant and autosomal recessive subtypes based on different patters of inheritance. Early mortality is associated with bone marrow failure, infections, lung and pulmonary complications as well as malignancy. The Igenomix Dyskeratosis Congenita Precision Panel can be used for an accurate and directed diagnosis as well as differential diagnosis of reticulate pigmentary disorders ultimately leading to a better management and prognosis of the disease. It provides a comprehensive analysis of the genes involved in this disease using next-generation sequencing (NGS) to fully understand the spectrum of relevant genes involved. Indications The Igenomix Dyskeratosis Congenita Precision Panel is used for patients with a clinical diagnosis or suspicion with or without the following symptoms: ‐ Abnormal skin pigmentation (tan-to-gray hyperpigmented or hypopigmented macules and patches) ‐ Nail dystrophy ‐ Skin atrophy and telangiectasia ‐ Alopecia of the skin, eyebrows and eyelashes ‐ Mucosal leukoplakia ‐ Bone marrow failure ‐ Dental manifestations Clinical Utility The clinical utility of this panel is: ‐ The genetic and molecular confirmation for an accurate clinical diagnosis of a symptomatic patient. 1 ‐ Early initiation of treatment involving a multidisciplinary team in the form of hematopoietic stem cell transplantation as well as medical care to prevent complications and early surveillance of malignancy. -

Joubert Syndrome Genereview
Title: Joubert Syndrome GeneReview — Molecular Genetics: Less Common Genetic Causes Authors: Parisi M, Glass I Updated: June 2017 Note: The following information is provided by the authors listed above and has not been reviewed by GeneReviews staff. Joubert Syndrome: Less Common Genetic Causes ARL13B B9D1 B9D2 CEP41 IFT172 KIF7 OFD1 (CXORF5) PDE6D POC1B TCTN1 TCTN3 TMEM138 TMEM231 TMEM237 (ALS2CR4) TTC21B ARL13B Gene structure. ARL13B is a ten-exon gene that encodes a 428-amino acid protein. Pathogenic variants. Two families with a phenotype typical of classic Joubert syndrome had missense and/or nonsense variants in this gene; one of these individuals also had evidence of a retinopathy [Cantagrel et al 2008]. Normal gene product. ARL13B encodes ADP-ribosylation factor-like protein 13B, a member of the ADP-ribosylation factor-like family. Multiple transcript variants result from alternate splicing; two protein isoforms are known. The AR13B protein is a small GTPase in the Ras superfamily that contains both N-terminal and C-terminal guanine nucleotide-binding motifs. It is localized to the cilia and plays a role in cilia formation and maintenance as well as sonic hedgehog signaling. Abnormal gene product. In C elegans, pathogenic variants in the homolog arl13 exhibit defective cilium morphology, localization, and anterograde intraflagellar transport [Cevik et al 2010]. Mice with defects in the murine ortholog have neural tube defects and polydactyly, as well as an embryonic-lethal phenotype [Cantagrel et al 2008, Doherty 2009]. B9D1. See Tables A and B. B9D2. See Tables A and B. CEP41 Gene structure. The gene consists of 11 exons and spans approximately 50 kb. -

Joubert Syndrome and Related Disorders
Brancati et al. Orphanet Journal of Rare Diseases 2010, 5:20 http://www.ojrd.com/content/5/1/20 REVIEW Open Access JoubertReview Syndrome and related disorders Francesco Brancati1,2, Bruno Dallapiccola3 and Enza Maria Valente*1,4 Abstract Joubert syndrome (JS) and related disorders (JSRD) are a group of developmental delay/multiple congenital anomalies syndromes in which the obligatory hallmark is the molar tooth sign (MTS), a complex midbrain-hindbrain malformation visible on brain imaging, first recognized in JS. Estimates of the incidence of JSRD range between 1/80,000 and 1/ 100,000 live births, although these figures may represent an underestimate. The neurological features of JSRD include hypotonia, ataxia, developmental delay, intellectual disability, abnormal eye movements, and neonatal breathing dysregulation. These may be associated with multiorgan involvement, mainly retinal dystrophy, nephronophthisis, hepatic fibrosis and polydactyly, with both inter- and intra-familial variability. JSRD are classified in six phenotypic subgroups: Pure JS; JS with ocular defect; JS with renal defect; JS with oculorenal defects; JS with hepatic defect; JS with orofaciodigital defects. With the exception of rare X-linked recessive cases, JSRD follow autosomal recessive inheritance and are genetically heterogeneous. Ten causative genes have been identified to date, all encoding for proteins of the primary cilium or the centrosome, making JSRD part of an expanding group of diseases called "ciliopathies". Mutational analysis of causative genes is available in few laboratories worldwide on a diagnostic or research basis. Differential diagnosis must consider in particular the other ciliopathies (such as nephronophthisis and Senior-Loken syndrome), distinct cerebellar and brainstem congenital defects and disorders with cerebro-oculo-renal manifestations. -

WES Gene Package Multiple Congenital Anomalie.Xlsx
Whole Exome Sequencing Gene package Multiple congenital anomalie, version 5, 1‐2‐2018 Technical information DNA was enriched using Agilent SureSelect Clinical Research Exome V2 capture and paired‐end sequenced on the Illumina platform (outsourced). The aim is to obtain 8.1 Giga base pairs per exome with a mapped fraction of 0.99. The average coverage of the exome is ~50x. Duplicate reads are excluded. Data are demultiplexed with bcl2fastq Conversion Software from Illumina. Reads are mapped to the genome using the BWA‐MEM algorithm (reference: http://bio‐bwa.sourceforge.net/). Variant detection is performed by the Genome Analysis Toolkit HaplotypeCaller (reference: http://www.broadinstitute.org/gatk/). The detected variants are filtered and annotated with Cartagenia software and classified with Alamut Visual. It is not excluded that pathogenic mutations are being missed using this technology. At this moment, there is not enough information about the sensitivity of this technique with respect to the detection of deletions and duplications of more than 5 nucleotides and of somatic mosaic mutations (all types of sequence changes). HGNC approved Phenotype description including OMIM phenotype ID(s) OMIM median depth % covered % covered % covered gene symbol gene ID >10x >20x >30x A4GALT [Blood group, P1Pk system, P(2) phenotype], 111400 607922 101 100 100 99 [Blood group, P1Pk system, p phenotype], 111400 NOR polyagglutination syndrome, 111400 AAAS Achalasia‐addisonianism‐alacrimia syndrome, 231550 605378 73 100 100 100 AAGAB Keratoderma, palmoplantar,