Rieck Et Al.- Supplementary Gene List Figure 1- FINAL
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

The Global Architecture Shaping the Heterogeneity and Tissue-Dependency of the MHC Class I Immunopeptidome Is Evolutionarily Conserved
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.28.317750; this version posted September 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The Global Architecture Shaping the Heterogeneity and Tissue-Dependency of the MHC Class I Immunopeptidome is Evolutionarily Conserved Authors Peter Kubiniok†1, Ana Marcu†2,3, Leon Bichmann†2,4, Leon Kuchenbecker4, Heiko Schuster1,5, David Hamelin1, Jérome Despault1, Kevin Kovalchik1, Laura Wessling1, Oliver Kohlbacher4,7,8,9,10 Stefan Stevanovic2,3,6, Hans-Georg Rammensee2,3,6, Marian C. Neidert11, Isabelle Sirois1, Etienne Caron1,12* Affiliations *Corresponding and Leading author: Etienne Caron ([email protected]) †Equal contribution to this work 1CHU Sainte-Justine Research Center, Montreal, QC H3T 1C5, Canada 2Department of Immunology, Interfaculty Institute for Cell Biology, University of Tübingen, Tübingen, Baden-Württemberg, 72076, Germany. 3Cluster of Excellence iFIT (EXC 2180) "Image-Guided and Functionally Instructed Tumor Therapies", University of Tübingen, Tübingen, Baden-Württemberg, 72076, Germany. 4Applied Bioinformatics, Dept. of Computer Science, University of Tübingen, Tübingen, Baden- Württemberg, 72074, Germany. 5Immatics Biotechnologies GmbH, Tübingen, 72076, Baden-Württemberg, Germany. 6DKFZ Partner Site Tübingen, German Cancer Consortium (DKTK), Tübingen, Baden- Württemberg, 72076, Germany. 7Institute for Bioinformatics and Medical Informatics, -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Tnfa-Induced Mucin 4 Expression Elicits Trastuzumab Resistance in HER2-Positive Breast Cancer María F
Published OnlineFirst October 3, 2016; DOI: 10.1158/1078-0432.CCR-16-0970 Cancer Therapy: Clinical Clinical Cancer Research TNFa-Induced Mucin 4 Expression Elicits Trastuzumab Resistance in HER2-Positive Breast Cancer María F. Mercogliano1, Mara De Martino1, Leandro Venturutti1, Martín A. Rivas2, Cecilia J. Proietti1, Gloria Inurrigarro3, Isabel Frahm3, Daniel H. Allemand4, Ernesto Gil Deza5, Sandra Ares5, Felipe G. Gercovich5, Pablo Guzman 6, Juan C. Roa6,7, Patricia V. Elizalde1, and Roxana Schillaci1 Abstract Purpose: Although trastuzumab administration improved the Results: TNFa overexpression turned trastuzumab-sensitive outcome of HER2-positive breast cancer patients, resistance cells and tumors into resistant ones. Histopathologic findings events hamper its clinical benefits. We demonstrated that TNFa revealed mucin foci in TNFa-producing tumors. TNFa induced stimulation in vitro induces trastuzumab resistance in HER2- upregulation of MUC4 that reduced trastuzumab binding to its positive breast cancer cell lines. Here, we explored the mechanism epitope and impaired ADCC. Silencing MUC4 enhanced trastu- of TNFa-induced trastuzumab resistance and the therapeutic zumab binding, increased ADCC, and overcame trastuzumab and strategies to overcome it. trastuzumab-emtansine antiproliferative effects in TNFa-overex- Experimental Design: Trastuzumab-sensitive breast cancer pressing cells. Accordingly, administration of TNFa-blocking cells, genetically engineered to stably overexpress TNFa,and antibodies downregulated MUC4 and sensitized de novo trastu- de novo trastuzumab-resistant tumors, were used to evaluate zumab-resistant breast cancer cells and tumors to trastuzumab. In trastuzumab response and TNFa-blocking antibodies effective- HER2-positive breast cancer samples, MUC4 expression was ness respectively. Immunohistochemistry and antibody-depen- found to be an independent predictor of poor disease-free survival dent cell cytotoxicity (ADCC), together with siRNA strategy, (P ¼ 0.008). -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

A Stealth Cloak for Cancer Cells
BMB Rep. 2021; 54(7): 344-355 BMB www.bmbreports.org Reports Invited Mini Review Mucin in cancer: a stealth cloak for cancer cells Dong-Han Wi1, Jong-Ho Cha2,3 & Youn-Sang Jung1,* 1Department of Life Science, Chung-Ang University, Seoul, 06974, 2Department of Biomedical Sciences, College of Medicine, Inha University, Incheon 22212, 3Department of Biomedical Science, Program in Biomedical Science and Engineering, Graduate school, Inha University, Incheon 22212, Korea Mucins are high molecular-weight epithelial glycoproteins and mucinous colorectal carcinoma (MCC) (3). Since tumor growth are implicated in many physiological processes, including epit- sites induce inhospitable conditions for them to survive, helial cell protection, signaling transduction, and tissue home- mucins are suggested as an oncogenic microenvironment that ostasis. Abnormality of mucus expression and structure contri- avoids hypoxia, acidic, and other biological hurdles. The com- butes to biological properties related to human cancer progress- position and structure of mucins enable them to mimic the ion. Tumor growth sites induce inhospitable conditions. Many surface of tumor cells like the surface of normal epithelial cells kinds of research suggest that mucins provide a microenviron- (4). Additionally, the mucus layer captures growth factors or ment to avoid hypoxia, acidic, and other biological conditions cytokines, contributing to cell growth of the tumor. Alter- that promote cancer progression. Given that the mucus layer natively, these properties interfere with the interaction bet- captures growth factors or cytokines, we propose that mucin ween the immune system and tumor cells. Indeed, a high helps to ameliorate inhospitable conditions in tumor-growing concentration of soluble mucins downregulates the motility sites. -

Role of Active Contraction and Tropomodulins in Regulating Actin Filament Length and Sarcomere Structure in Developing Zebrafish Skeletal Muscle
ORIGINAL RESEARCH published: 31 March 2016 doi: 10.3389/fphys.2016.00091 Role of Active Contraction and Tropomodulins in Regulating Actin Filament Length and Sarcomere Structure in Developing Zebrafish Skeletal Muscle Lise Mazelet 1, Matthew O. Parker 2, Mei Li 3, Anders Arner 3 and Rachel Ashworth 4* 1 School of Biological and Chemical Sciences, Queen Mary, University of London, London, UK, 2 School of Health Sciences and Social Work, University of Portsmouth, Portsmouth, UK, 3 Department of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden, 4 The Blizard Institute/Institute of Health Sciences Education, Barts and The London School of Medicine and Dentistry, London, UK Whilst it is recognized that contraction plays an important part in maintaining the structure and function of mature skeletal muscle, its role during development remains undefined. In this study the role of movement in skeletal muscle maturation was investigated in intact zebrafish embryos using a combination of genetic and pharmacological approaches. An immotile mutant line (cacnb1ts25) which lacks functional voltage-gated calcium channels (dihydropyridine receptors) in the muscle and pharmacological immobilization of embryos Edited by: with a reversible anesthetic (Tricaine), allowed the study of paralysis (in mutants and Catherine Coirault, anesthetized fish) and recovery of movement (reversal of anesthetic treatment). The Institut National de la Santé et de la Recherche Médicale, France effect of paralysis in early embryos (aged between 17 and 24 hours post-fertilization, hpf) Reviewed by: on skeletal muscle structure at both myofibrillar and myofilament level was determined Corrado Poggesi, using both immunostaining with confocal microscopy and small angle X-ray diffraction. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
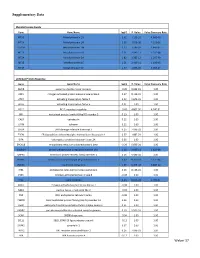
Weber 37 Supplementary Data
Supplementary Data Metallothionein Family Gene Gene Name logFC P. Value False Discovery Rate MT1G Metallothionein 1G 2.62 1.19E-10 4.66E-08 MT1H Metallothionein 1H 2.58 4.87E-08 7.12E-06 MT1M Metallothionein 1M 2.23 1.18E-07 1.44E-05 MT1X Metallothionein 1X 2.21 2.64E-11 1.25E-08 MT2A Metallothionein 2A 1.82 1.38E-12 1.26E-09 MT1E Metallothionein 1E 1.51 2.41E-11 1.18E-08 MT1F Metallothionein 1F 1.47 1.85E-09 4.09E-07 Unfolded Protein Response Gene Gene Name logFC P. Value False Discovery Rate AMFR autocrine motility factor receptor -0.20 9.94E-01 1.00 ASK1 mitogen-activated protein kinase kinase kinase 5 0.27 8.11E-01 1.00 ATF4 activating transcription factor 4 0.22 9.67E-01 1.00 ATF6 activating transcription factor 6 0.01 1.00 1.00 BCL2 BCL2, apoptosis regulator -0.43 4.80E-02 4.24E-01 BIP heat shock protein family A (Hsp70) member 5 0.13 1.00 1.00 CALR calreticulin 0.12 1.00 1.00 CANX calnexin 0.12 1.00 1.00 CHOP DNA damage inducible transcript 3 0.25 7.99E-01 1.00 EDEM ER degradation enhancing alpha-mannosidase like protein 1 0.37 1.88E-01 1.00 EIFA eukaryotic translation initiation factor 2A 0.06 1.00 1.00 ERO1LB endoplasmic reticulum oxidoreductase 1 beta -0.29 5.69E-01 1.00 GADD34 protein phosphatase 1 regulatory subunit 15A 1.18 3.66E-11 1.62E-08 GRP94 heat shock protein 90 beta family member 1 0.16 9.98E-01 1.00 HSPH1 heat shock protein family H (Hsp110) member 1 0.62 9.70E-06 5.12E-04 INSIG1 insulin induced gene 1 1.69 8.47E-18 3.88E-14 IRE1 endoplasmic reticulum to nucleus signaling 1 0.26 8.13E-01 1.00 JNK1 mitogen-activated