Molecular Systematics of the Genus Uvularia and Selected Liliales Based Upon Matk and Rbcl Gene Sequence Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Asparagus Densiflorus SCORE: 15.0 RATING: High Risk (Kunth) Jessop
TAXON: Asparagus densiflorus SCORE: 15.0 RATING: High Risk (Kunth) Jessop Taxon: Asparagus densiflorus (Kunth) Jessop Family: Asparagaceae Common Name(s): asparagus fern Synonym(s): Asparagopsis densiflora Kunth foxtail fern Asparagus myriocladus Baker plume asparagus Protasparagus densiflorus (Kunth) Oberm. regal fern Sprenger's asparagus fern Assessor: Chuck Chimera Status: Assessor Approved End Date: 16 Feb 2021 WRA Score: 15.0 Designation: H(HPWRA) Rating: High Risk Keywords: Tuberous Geophyte, Naturalized, Environmental Weed, Dense Cover, Bird-Dispersed Qsn # Question Answer Option Answer 101 Is the species highly domesticated? y=-3, n=0 n 102 Has the species become naturalized where grown? 103 Does the species have weedy races? Species suited to tropical or subtropical climate(s) - If 201 island is primarily wet habitat, then substitute "wet (0-low; 1-intermediate; 2-high) (See Appendix 2) High tropical" for "tropical or subtropical" 202 Quality of climate match data (0-low; 1-intermediate; 2-high) (See Appendix 2) High 203 Broad climate suitability (environmental versatility) y=1, n=0 n Native or naturalized in regions with tropical or 204 y=1, n=0 y subtropical climates Does the species have a history of repeated introductions 205 y=-2, ?=-1, n=0 y outside its natural range? 301 Naturalized beyond native range y = 1*multiplier (see Appendix 2), n= question 205 y 302 Garden/amenity/disturbance weed n=0, y = 1*multiplier (see Appendix 2) n 303 Agricultural/forestry/horticultural weed n=0, y = 2*multiplier (see Appendix 2) n 304 -
Calochortus Flexuosus S. Watson (Winding Mariposa Lily): a Technical Conservation Assessment
Calochortus flexuosus S. Watson (winding mariposa lily): A Technical Conservation Assessment Prepared for the USDA Forest Service, Rocky Mountain Region, Species Conservation Project July 24, 2006 Susan Spackman Panjabi and David G. Anderson Colorado Natural Heritage Program Colorado State University Fort Collins, CO Peer Review Administered by Center for Plant Conservation Panjabi, S.S. and D.G. Anderson. (2006, July 24). Calochortus flexuosus S. Watson (winding mariposa lily): a technical conservation assessment. [Online]. USDA Forest Service, Rocky Mountain Region. Available: http://www.fs.fed.us/r2/projects/scp/assessments/calochortusflexuosus.pdf [date of access]. ACKNOWLEDGMENTS This research was facilitated by the helpfulness and generosity of many experts, particularly Leslie Stewart, Peggy Fiedler, Marilyn Colyer, Peggy Lyon, Lynn Moore, and William Jennings. Their interest in the project and time spent answering questions were extremely valuable, and their insights into the distribution, habitat, and ecology of Calochortus flexuosus were crucial to this project. Thanks also to Greg Hayward, Gary Patton, Jim Maxwell, Andy Kratz, and Joy Bartlett for assisting with questions and project management. Thanks to Kimberly Nguyen for her work on the layout and for bringing this assessment to Web publication. Jane Nusbaum and Barbara Brayfield provided crucial financial oversight. Peggy Lyon and Marilyn Colyer provided valuable insights based on their experiences with C. flexuosus. Leslie Stewart provided information specific to the San Juan Resource Area of the Bureau of Land Management, including the Canyons of the Ancients National Monument. Annette Miller provided information on C. flexuosusseed storage status. Drs. Ron Hartman and Ernie Nelson provided access to specimens of C. -

Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE
Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE LILIACEAE de Jussieu 1789 (Lily Family) (also see AGAVACEAE, ALLIACEAE, ALSTROEMERIACEAE, AMARYLLIDACEAE, ASPARAGACEAE, COLCHICACEAE, HEMEROCALLIDACEAE, HOSTACEAE, HYACINTHACEAE, HYPOXIDACEAE, MELANTHIACEAE, NARTHECIACEAE, RUSCACEAE, SMILACACEAE, THEMIDACEAE, TOFIELDIACEAE) As here interpreted narrowly, the Liliaceae constitutes about 11 genera and 550 species, of the Northern Hemisphere. There has been much recent investigation and re-interpretation of evidence regarding the upper-level taxonomy of the Liliales, with strong suggestions that the broad Liliaceae recognized by Cronquist (1981) is artificial and polyphyletic. Cronquist (1993) himself concurs, at least to a degree: "we still await a comprehensive reorganization of the lilies into several families more comparable to other recognized families of angiosperms." Dahlgren & Clifford (1982) and Dahlgren, Clifford, & Yeo (1985) synthesized an early phase in the modern revolution of monocot taxonomy. Since then, additional research, especially molecular (Duvall et al. 1993, Chase et al. 1993, Bogler & Simpson 1995, and many others), has strongly validated the general lines (and many details) of Dahlgren's arrangement. The most recent synthesis (Kubitzki 1998a) is followed as the basis for familial and generic taxonomy of the lilies and their relatives (see summary below). References: Angiosperm Phylogeny Group (1998, 2003); Tamura in Kubitzki (1998a). Our “liliaceous” genera (members of orders placed in the Lilianae) are therefore divided as shown below, largely following Kubitzki (1998a) and some more recent molecular analyses. ALISMATALES TOFIELDIACEAE: Pleea, Tofieldia. LILIALES ALSTROEMERIACEAE: Alstroemeria COLCHICACEAE: Colchicum, Uvularia. LILIACEAE: Clintonia, Erythronium, Lilium, Medeola, Prosartes, Streptopus, Tricyrtis, Tulipa. MELANTHIACEAE: Amianthium, Anticlea, Chamaelirium, Helonias, Melanthium, Schoenocaulon, Stenanthium, Veratrum, Toxicoscordion, Trillium, Xerophyllum, Zigadenus. -

Cally Plant List a ACIPHYLLA Horrida
Cally Plant List A ACIPHYLLA horrida ACONITUM albo-violaceum albiflorum ABELIOPHYLLUM distichum ACONITUM cultivar ABUTILON vitifolium ‘Album’ ACONITUM pubiceps ‘Blue Form’ ACAENA magellanica ACONITUM pubiceps ‘White Form’ ACAENA species ACONITUM ‘Spark’s Variety’ ACAENA microphylla ‘Kupferteppich’ ACONITUM cammarum ‘Bicolor’ ACANTHUS mollis Latifolius ACONITUM cammarum ‘Franz Marc’ ACANTHUS spinosus Spinosissimus ACONITUM lycoctonum vulparia ACANTHUS ‘Summer Beauty’ ACONITUM variegatum ACANTHUS dioscoridis perringii ACONITUM alboviolaceum ACANTHUS dioscoridis ACONITUM lycoctonum neapolitanum ACANTHUS spinosus ACONITUM paniculatum ACANTHUS hungaricus ACONITUM species ex. China (Ron 291) ACANTHUS mollis ‘Long Spike’ ACONITUM japonicum ACANTHUS mollis free-flowering ACONITUM species Ex. Japan ACANTHUS mollis ‘Turkish Form’ ACONITUM episcopale ACANTHUS mollis ‘Hollard’s Gold’ ACONITUM ex. Russia ACANTHUS syriacus ACONITUM carmichaelii ‘Spätlese’ ACER japonicum ‘Aconitifolium’ ACONITUM yezoense ACER palmatum ‘Filigree’ ACONITUM carmichaelii ‘Barker’s Variety’ ACHILLEA grandifolia ACONITUM ‘Newry Blue’ ACHILLEA ptarmica ‘Perry’s White’ ACONITUM napellus ‘Bergfürst’ ACHILLEA clypeolata ACONITUM unciniatum ACIPHYLLA monroi ACONITUM napellus ‘Blue Valley’ ACIPHYLLA squarrosa ACONITUM lycoctonum ‘Russian Yellow’ ACIPHYLLA subflabellata ACONITUM japonicum subcuneatum ACONITUM meta-japonicum ADENOPHORA aurita ACONITUM napellus ‘Carneum’ ADIANTUM aleuticum ‘Japonicum’ ACONITUM arcuatum B&SWJ 774 ADIANTUM aleuticum ‘Miss Sharples’ ACORUS calamus ‘Argenteostriatus’ -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Introduction to Common Native & Invasive Freshwater Plants in Alaska
Introduction to Common Native & Potential Invasive Freshwater Plants in Alaska Cover photographs by (top to bottom, left to right): Tara Chestnut/Hannah E. Anderson, Jamie Fenneman, Vanessa Morgan, Dana Visalli, Jamie Fenneman, Lynda K. Moore and Denny Lassuy. Introduction to Common Native & Potential Invasive Freshwater Plants in Alaska This document is based on An Aquatic Plant Identification Manual for Washington’s Freshwater Plants, which was modified with permission from the Washington State Department of Ecology, by the Center for Lakes and Reservoirs at Portland State University for Alaska Department of Fish and Game US Fish & Wildlife Service - Coastal Program US Fish & Wildlife Service - Aquatic Invasive Species Program December 2009 TABLE OF CONTENTS TABLE OF CONTENTS Acknowledgments ............................................................................ x Introduction Overview ............................................................................. xvi How to Use This Manual .................................................... xvi Categories of Special Interest Imperiled, Rare and Uncommon Aquatic Species ..................... xx Indigenous Peoples Use of Aquatic Plants .............................. xxi Invasive Aquatic Plants Impacts ................................................................................. xxi Vectors ................................................................................. xxii Prevention Tips .................................................... xxii Early Detection and Reporting -

Taxonomy, Morphology and Palynology of Aegilops Vavilovii (Zhuk.) Chennav
African Journal of Agricultural Research Vol. 5(20), pp. 2841-2849, 18 October, 2010 Available online at http://www.academicjournals.org/AJAR ISSN 1991-637X ©2010 Academic Journals Full Length Research Paper Taxonomy, morphology and palynology of Aegilops vavilovii (Zhuk.) Chennav. (Poaceae: Triticeae) Evren Cabi1* Musa Doan1 Hülya Özler2, Galip Akaydin3 and Alptekin Karagöz4 1Department of Biological Sciences, Faculty of Arts and Sciences, Middle East Technical University, Ankara, Turkey. 2Department of Biology, Faculty of Arts and Sciences, Sinop University, Sinop Turkey. 3Department of Biology Education, Hacettepe University, 06800 Ankara, Turkey. 4Department of Biology, Aksaray University, Aksaray, Turkey. Accepted 23 September, 2010 Aegilops vavilovii (Zhuk.) Chennav., a rare species, was collected from Southeast Anatolia, Turkey. During the field studies of the project “Taxonomic revision of Tribe Triticeae in Turkey”, Ae. vavilovii was accidentally recollected from three localities in anliurfa and Mardin provinces in 2007 and 2008, respectively. The main objective of this study is to shed light on the diagnostic characteristics of this rare species including its morphological, palynological and micro morphological features. Moreover, an emended and expanded description, distribution, phenology and ecology of this rare species are also provided. A. vavilovii and A. crassa are naturally found in the Southeastern part of Turkey and they share similar morphological features that caused a confused taxonomy. Pollen grains of A. vavilovii are heteropolar, monoporate and spheroidal (A/B: 1,13) typically as Poaceous. However, it generally, prefers clayish loam soils that are slightly alkaline (pH 7.7) with low organic content (1.54%). Although it is a rare species with very narrow area of distribution, very few samples have been represented in ex situ collections and the species has not been involved in any in situ conservation activities to save its genetic resources in Turkey. -

Tracing History
Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 911 Tracing History Phylogenetic, Taxonomic, and Biogeographic Research in the Colchicum Family BY ANNIKA VINNERSTEN ACTA UNIVERSITATIS UPSALIENSIS UPPSALA 2003 Dissertation presented at Uppsala University to be publicly examined in Lindahlsalen, EBC, Uppsala, Friday, December 12, 2003 at 10:00 for the degree of Doctor of Philosophy. The examination will be conducted in English. Abstract Vinnersten, A. 2003. Tracing History. Phylogenetic, Taxonomic and Biogeographic Research in the Colchicum Family. Acta Universitatis Upsaliensis. Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 911. 33 pp. Uppsala. ISBN 91-554-5814-9 This thesis concerns the history and the intrafamilial delimitations of the plant family Colchicaceae. A phylogeny of 73 taxa representing all genera of Colchicaceae, except the monotypic Kuntheria, is presented. The molecular analysis based on three plastid regions—the rps16 intron, the atpB- rbcL intergenic spacer, and the trnL-F region—reveal the intrafamilial classification to be in need of revision. The two tribes Iphigenieae and Uvularieae are demonstrated to be paraphyletic. The well-known genus Colchicum is shown to be nested within Androcymbium, Onixotis constitutes a grade between Neodregea and Wurmbea, and Gloriosa is intermixed with species of Littonia. Two new tribes are described, Burchardieae and Tripladenieae, and the two tribes Colchiceae and Uvularieae are emended, leaving four tribes in the family. At generic level new combinations are made in Wurmbea and Gloriosa in order to render them monophyletic. The genus Androcymbium is paraphyletic in relation to Colchicum and the latter genus is therefore expanded. -

Evaluation of Agro-Morphological Diversity in Wild Relatives of Wheat Collected in Iran
J. Agr. Sci. Tech. (2017) Vol. 19: 943-956 Evaluation of Agro-Morphological Diversity in Wild Relatives of Wheat Collected in Iran A. Pour-Aboughadareh1*, J. Ahmadi1, A. A. Mehrabi2, M. Moghaddam3, and A. Etminan4 ABSTRACT In this study, a core collection of 180 Aegilops and Triticum accessions belonging to six diploid (T. boeoticum Bioss., T. urartu Gandilyan., Ae. speltoides Tausch., Ae. tauschii Coss., Ae. caudata L. and Ae. umbellulata Zhuk.), five tetraploid (T. durum, Ae. neglecta Req. ex Bertol., Ae. cylindrica Host. and Ae. crassa Boiss) and one hexaploid (T. aestivum L.) species collected from different regions of Iran were evaluated using 20 agro- morphological characters. Statistical analysis showed significant differences among accessions. The Shannon-Weaver (HʹSW) and Nei’s (HʹN) genetic diversity indices disclosed intermediate to high diversity for most characters in both Aegilops and Triticum core sets. In factor analysis, the first five components justified 82.17% of the total of agro- morphological variation. Based on measured characters, the 180 accessions were separated into two major groups by cluster analysis. Furthermore, based on the 2D-plot generated using two discriminant functions, different species were separated into six groups, so that distribution of species accorded with their genome construction. Overall, our results revealed considerable levels of genetic diversity among studied Iranian Aegilops and Triticum accessions, which can open up new avenues for rethinking the connections between wild relatives to explore valuable agronomic traits for the improvement and adaptation of wheat. Keywords: Aegilops, Multivariate analysis, Phenotypic diversity, Triticum. INTRODUCTION traits, such as disease- or pest-resistance and even yield improvement. -
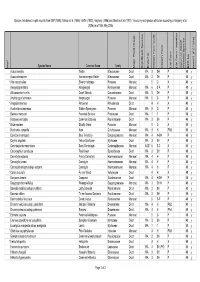
BFS048 Site Species List
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

Nordic Journal of Botany NJB-02477 She, R., Zhao, P
Nordic Journal of Botany NJB-02477 She, R., Zhao, P. , Zhou, H., Yue, M., Yan, F., Hu, G., Gao, X. and Zhang, S. 2020. Complete chloroplast genomes of Liliaceae (s.l.) species: comparative genomic and phylogenetic analyses. - Nordic Journal of Botany 2019: e02477 Appendix 1 Table A1. List of taxa sampled in this study and species accessions numbers (GenBank). Genus Species Accession number Genus Species Accession number Aletris fauriei KT898912 Polygonatum sibiricum KT695605 Aletris Polygonatum Aletris spicata KT898911 Polygonatum verticillatum KT722981 Allium cepa KM088013 Lilium amabile KY940845 Allium obliquum MG670111 Lilium bakerianum KY748301 Allium Allium prattii MG739457 Lilium brownii KY748296 Allium victorialis MF687749 Lilium bulbiferum MG574829 Amana anhuiensis KY401423 Lilium callosum KY940846 Amana edulis KY401425 Lilium cernuum KX354692 Amana Amana erythronioides KY401424 Lilium distichum KT376489 Amana kuocangshanica KY401426 Lilium duchartrei KY748300 Amana wanzhensis KY401422 Lilium fargesii KX592156 Fritillaria cirrhosa KF769143 Lilium hansonii KM103364 Lilium Fritillaria eduardii MF947708 Lilium henryi KY748302 Fritillaria hupehensis KF712486 Lilium lancifolium KY748297 Fritillaria meleagroides MF947710 Lilium leucanthum KY748299 Fritillaria persica MF947709 Lilium longiflorum KC968977 Fritillaria Fritillaria thunbergii KY646165 Lilium philadelphicum KY940847 Fritillaria unibracteata var. KF769142 Lilium primulinum var. ochraceum KY748298 wabuensis Fritillaria ussuriensis KY646166 Lilium taliense KY009938 Fritillaria -

Biodiversity and Ecology of Critically Endangered, Rûens Silcrete Renosterveld in the Buffeljagsrivier Area, Swellendam
Biodiversity and Ecology of Critically Endangered, Rûens Silcrete Renosterveld in the Buffeljagsrivier area, Swellendam by Johannes Philippus Groenewald Thesis presented in fulfilment of the requirements for the degree of Masters in Science in Conservation Ecology in the Faculty of AgriSciences at Stellenbosch University Supervisor: Prof. Michael J. Samways Co-supervisor: Dr. Ruan Veldtman December 2014 Stellenbosch University http://scholar.sun.ac.za Declaration I hereby declare that the work contained in this thesis, for the degree of Master of Science in Conservation Ecology, is my own work that have not been previously published in full or in part at any other University. All work that are not my own, are acknowledge in the thesis. ___________________ Date: ____________ Groenewald J.P. Copyright © 2014 Stellenbosch University All rights reserved ii Stellenbosch University http://scholar.sun.ac.za Acknowledgements Firstly I want to thank my supervisor Prof. M. J. Samways for his guidance and patience through the years and my co-supervisor Dr. R. Veldtman for his help the past few years. This project would not have been possible without the help of Prof. H. Geertsema, who helped me with the identification of the Lepidoptera and other insect caught in the study area. Also want to thank Dr. K. Oberlander for the help with the identification of the Oxalis species found in the study area and Flora Cameron from CREW with the identification of some of the special plants growing in the area. I further express my gratitude to Dr. Odette Curtis from the Overberg Renosterveld Project, who helped with the identification of the rare species found in the study area as well as information about grazing and burning of Renosterveld.