Population Genetics for Autosomal STR Loci in Sikh Population Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Any Person May Make a Complaint About The
HIGH COURT OF CHHATTISGARH AT BILASPUR WRIT PETITION (C) No. 4944 of 2009 PETITIONER : Rajendra Singh (Arora). V E R S U S RESPONDENTS : State of Chhattisgarh & Others. PETITION UNDER ARTICLE 226/227 OF THE CONSTITUTION OF INDIA SB: Hon’ble Shri Satish K. Agnihotri, J. --------------------------------------------------------------------------------------------- Present: Shri Kishore Shrivastava, Senior Advocate with Shri Sanjay Tamrakar, Shri Ashish Shirvastava and Shri Anshuman Shrivastava, Advocates for the petitioner. Shri V.V.S.Moorthy, Deputy Advocate General for the State/ respondent No. 1, 2, 4 and 5. Dr. N.K.Shukla, Senior Advocate with Shri Aditya Khare, Advocates for the respondent No. 7 and 8. --------------------------------------------------------------------------------------------- O R D E R (Delivered on 10th day of July, 2013) 1. The petitioner mainly challenges the order dated 23.07.2007 (Annexure P/14) passed by the High Power State Level Caste Scrutiny Committee i.e. respondent No. 3 (for short ‘the Committee’) wherein the petitioner has been found that he does not belong to “Lohar” i.e. Other Backward Class category, and also the subsequent order dated 28.08.2009 (Annexure P/2) wherein on the basis of the order dated 22.09.2008 (Annexure P/1) the petitioner was disqualified for a further period of five years in view of the provisions of section 19(2) of the Chhattisgarh Municipal Corporation Act, 1956 (for short 'the Act, 1956'). 2. The facts, in brief, are that the petitioner, in the year 2000, contested the election as Councillor from Ward No. 22, Municipal 2 Corporation, Bhilai, declaring himself as a member of “Lohar” community that comes within OBC category on the basis of social status certificate dated 10.04.2000 (Annexure P/3). -

Punjab Public Works Department (B&R)
Punjab Public Works Department (B&R) Establishment Chart ( Dated : 17.09.2021 ) Chief Engineer (Civil) S. Name of Officer/ Email Qualification Present Place of Posting Date of Home Date of No address/ Mobile No. Posting District Birth 1. Er. Arun Kumar M.E. Chief Engineer (North) 12.11.2018 Ludhiana 28.11.1964 [email protected] Incharge of:- [email protected] Construction Circle, Amritsar 9872253744 and Hoshiarpur from 08.03.2019 And Additional Charge Chief Engineer (Headquarter-1), and Chief Engineer (Headquarter-2) and Nodal Officer (Punjab Vidhan Sabha Matters)(Plan Roads) 2. Er. Amardeep Singh Brar, B.E.(Civil) Chief Engineer (West) 03.11.2020 Faridkot 25.03.1965 Chief Engineer, Incharge of: [email protected] Construction Circle Bathinda, and 9915400934 Ferozepur 3. Er.N.R.Goyal, Chief Engineer (South) 03.11.2020 Fazilka 15.05.1964 Chief Engineer Incharge of: [email protected] Construction Circle Patiala - 1 and [email protected] Sangrur, Nodal Officer –Link [email protected] Roads,PMGSY & NABARD 9356717117 Additional Charge Chief Engineer (Quality Assurance) from 19.04.2021 & Chief Vigilance Officer of PWD (B&R) Chief Engineer (NH) from 20.08.2021 Incharge of: National Highway Circle Amritsar, 4. Er.B.S.Tuli, M.E.(Irrigation) ChiefChandigarh, Engineer Fe (Centrozepurral) and Ludhiana 03.11.2020 Ludhiana 15.09.1964 Chief Engineer and Hydraulic Incha rge of: [email protected] Structure) Construction Circle No. 1 & 2 Jalandhar., 9814183304 Construction Circle Pathankot. Nodal Officer (Railways) from 03.11.2020 , Jang-e-Azadi Memorial, Kartarpur and Works under 3054 & 5054 Head 5. -

The Sikh Dilemma: the Partition of Punjab 1947
The Sikh Dilemma: The Partition of Punjab 1947 Busharat Elahi Jamil Abstract The Partition of India 1947 resulted in the Partition of the Punjab into two, East and West. The 3rd June Plan gave a sense of uneasiness and generated the division of dilemma among the large communities of the British Punjab like Muslims, Hindus and Sikh besetting a holocaust. This situation was beneficial for the British and the Congress. The Sikh community with the support of Congress wanted the proportion of the Punjab according to their own violation by using different modules of deeds. On the other hand, for Muslims the largest populous group of the Punjab, by using the platform of Muslim League showed the resentment because they wanted the decision on the Punjab according to their requirements. Consequently the conflict caused the world’s bloodiest partition and the largest migration of the history. Introduction The Sikhs were the third largest community of the United Punjab before India’s partition. The Sikhs had the historic religious, economic and socio-political roots in the Punjab. Since the annexation of the Punjab, they were faithful with the British rulers and had an influence in the Punjabi society, even enjoying various privileges. But in the 20th century, the Muslims 90 Pakistan Vision Vol. 17 No. 1 Independence Movement in India was not only going to divide the Punjab but also causing the division of the Sikh community between East and West Punjab, which confused the Sikh leadership. So according to the political scenarios in different timings, Sikh leadership changed their demands and started to present different solutions of the Sikh enigma for the geographical transformation of the province. -

Changing Caste Relations and Emerging Contestations in Punjab
CHANGING CASTE RELATIONS AND EMERGING CONTESTATIONS IN PUNJAB PARAMJIT S. JUDGE When scholars and political leaders characterised Indian society as unity in diversity, there were simultaneous efforts in imagining India as a civilisational unity also. The consequences of this ‘imagination’ are before us in the form of the emergence of religious nationalism that ultimately culminated into the partition of the country. Why have I started my discussion with the issue of religious nationalism and partition? The reason is simple. Once we assume that a society like India could be characterised in terms of one caste hierarchical system, we are essentially constructing the discourse of dominant Hindu civilisational unity. Unlike class and gender hierarchies which are exist on economic and sexual bases respectively, all castes cannot be aggregated and arranged in hierarchy along one axis. Any attempt at doing so would amount to the construction of India as essentially the Hindu India. Added to this issue is the second dimension of hierarchy, which could be seen by separating Varna from caste. Srinivas (1977) points out that Varna is fixed, whereas caste is dynamic. Numerous castes comprise each Varna, the exception to which is the Brahmin caste whose caste differences remain within the caste and are unknown to others. We hardly know how to distinguish among different castes of Brahmins, because there is complete absence of knowledge about various castes among them. On the other hand, there is detailed information available about all the scheduled castes and backward classes. In other words, knowledge about castes and their place in the stratification system is pre- determined by the enumerating agency. -

2021 Daily Prayer Guide for All People Groups & Unreached People
2021 Daily Prayer Guide for all People Groups & Unreached People Groups = LR-UPGs = of South Asia Joshua Project data, www.joshuaproject.net (India DPG is separate) AGWM Western edition I give credit & thanks to Create International for permission to use their PG photos. 2021 Daily Prayer Guide for all People Groups & LR-UPGs = Least-Reached-Unreached People Groups of South Asia = this DPG SOUTH ASIA SUMMARY: 873 total People Groups; 733 UPGs The 6 countries of South Asia (India; Bangladesh; Nepal; Sri Lanka; Bhutan; Maldives) has 3,178 UPGs = 42.89% of the world's total UPGs! We must pray and reach them! India: 2,717 total PG; 2,445 UPGs; (India is reported in separate Daily Prayer Guide) Bangladesh: 331 total PG; 299 UPGs; Nepal: 285 total PG; 275 UPG Sri Lanka: 174 total PG; 79 UPGs; Bhutan: 76 total PG; 73 UPGs; Maldives: 7 total PG; 7 UPGs. Downloaded from www.joshuaproject.net in September 2020 LR-UPG definition: 2% or less Evangelical & 5% or less Christian Frontier (FR) definition: 0% to 0.1% Christian Why pray--God loves lost: world UPGs = 7,407; Frontier = 5,042. Color code: green = begin new area; blue = begin new country "Prayer is not the only thing we can can do, but it is the most important thing we can do!" Luke 10:2, Jesus told them, "The harvest is plentiful, but the workers are few. Ask the Lord of the harvest, therefore, to send out workers into his harvest field." Why Should We Pray For Unreached People Groups? * Missions & salvation of all people is God's plan, God's will, God's heart, God's dream, Gen. -

2021 Daily Prayer Calendar
2021 Daily Prayer Calendar of world's largest Unreached People Groups AGWM Western edition 2021 Daily Prayer Calendar of world's largest Unreached People Groups Joshua Project data downloaded October 2020 = www.joshuaproject.net LR-UPG definition: less than 2% Evangelical & less than 5% Christian; world UPG = 7,407 Frontier (FR) definition: 0% to 0.1% Christian; world Frontier = 5,042 (2-2021) 2021 = 1 PG photo = date (no PG photo avail. = country flag) Explanation Bangladesh = country of Shaikh = people group name 135,226,000 = people group population daily Bengali = main language data: Islam = main religion LR-UPG or Frontier = Least-Reached-Unreached People Group Let's dream God's dreams & fulfill God's visions -- God dreams of all people groups knowing & loving Him! * * * Revelation 7:9, "After this I looked and there before me was a great multitude that no one could count, from every nation, tribe, people and language, standing before the throne and in front of the Lamb." Why Should We Pray For Unreached People Groups? * Missions & salvation of all people is God's plan, God's will, God's heart, God's dream, Gen. 3:15! * In the Great Commissions Jesus commands us to reach all peoples in the world, Matt. 28:19-20! * People without Jesus are eternally lost, & Jesus is the only One who can save them, John 14:6! * We have been given "the ministry & message of reconciliation", in Christ, 2 Cor. 5:18-20! * Jesus’ return is intimately tied to reaching all people groups, Matt. 24:14; Rev. 7:9! * Jesus commanded us to pray to Him to send workers -

Displacement and Development: Long Term Impacts of Population Transfer in India ∗
Displacement and Development: Long Term Impacts of Population Transfer in India ∗ Prashant Bharadwajy Rinchan Ali Mirzaz August 2018 Abstract The partition of British India in 1947 resulted in one of the largest and most rapid migrations and population transfers of the 20th century. Using refugee pres- ence by 1951 as a measure for the intensity of the impact of the population transfer, and district level data on agricultural output between 1911-2009 from India, we find using difference in differences and event study approaches that areas that received more refugees have higher average yields, are more likely to take up high yielding varieties of seeds, and are more likely to use agricultural technologies. The increase in yields and use of agricultural technology coincide with the timing of the Green Revolution in India. Using pre-partition data, we show that refugee placement is uncorrelated with soil and water table characteristics, agricultural infrastructure, and agricultural yields prior to 1947; hence, the effects are not explained by selec- tive movement into districts with a higher potential for agricultural development. We highlight refugee literacy and land reforms in areas with refugees as two of the many potential mechanisms that could be driving these effects. JEL Codes: O13, O33, O15, Q16, N55 ∗This project has benefitted from comments by Latika Chaudhary, James Fenske, Bishnupriya Gupta, Asim Khwaja, Takashi Kurosaki, Atif Mian, participants of the 17th World Economic History Congress, organizers and participants of the 10th International Conference on Migration and Development, and various seminar participants. yUniversity of California San Diego & NBER. Corresponding author: Economics Dept, 9500 Gilman Dr. -
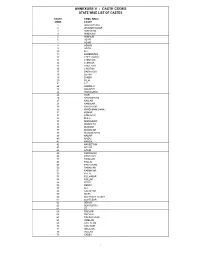
Annexure V - Caste Codes State Wise List of Castes
ANNEXURE V - CASTE CODES STATE WISE LIST OF CASTES STATE TAMIL NADU CODE CASTE 1 ADDI DIRVISA 2 AKAMOW DOOR 3 AMBACAM 4 AMBALAM 5 AMBALM 6 ASARI 7 ASARI 8 ASOOY 9 ASRAI 10 B.C. 11 BARBER/NAI 12 CHEETAMDR 13 CHELTIAN 14 CHETIAR 15 CHETTIAR 16 CRISTAN 17 DADA ACHI 18 DEYAR 19 DHOBY 20 DILAI 21 F.C. 22 GOMOLU 23 GOUNDEL 24 HARIAGENS 25 IYAR 26 KADAMBRAM 27 KALLAR 28 KAMALAR 29 KANDYADR 30 KIRISHMAM VAHAJ 31 KONAR 32 KONAVAR 33 M.B.C. 34 MANIGAICR 35 MOOPPAR 36 MUDDIM 37 MUNALIAR 38 MUSLIM/SAYD 39 NADAR 40 NAIDU 41 NANDA 42 NAVEETHM 43 NAYAR 44 OTHEI 45 PADAIACHI 46 PADAYCHI 47 PAINGAM 48 PALLAI 49 PANTARAM 50 PARAIYAR 51 PARMYIAR 52 PILLAI 53 PILLAIMOR 54 POLLAR 55 PR/SC 56 REDDY 57 S.C. 58 SACHIYAR 59 SC/PL 60 SCHEDULE CASTE 61 SCHTLEAR 62 SERVA 63 SOWRSTRA 64 ST 65 THEVAR 66 THEVAR 67 TSHIMA MIAR 68 UMBLAR 69 VALLALAM 70 VAN NAIR 71 VELALAR 72 VELLAR 73 YADEV 1 STATE WISE LIST OF CASTES STATE MADHYA PRADESH CODE CASTE 1 ADIWARI 2 AHIR 3 ANJARI 4 BABA 5 BADAI (KHATI, CARPENTER) 6 BAMAM 7 BANGALI 8 BANIA 9 BANJARA 10 BANJI 11 BASADE 12 BASOD 13 BHAINA 14 BHARUD 15 BHIL 16 BHUNJWA 17 BRAHMIN 18 CHAMAN 19 CHAWHAN 20 CHIPA 21 DARJI (TAILOR) 22 DHANVAR 23 DHIMER 24 DHOBI 25 DHOBI (WASHERMAN) 26 GADA 27 GADARIA 28 GAHATRA 29 GARA 30 GOAD 31 GUJAR 32 GUPTA 33 GUVATI 34 HARJAN 35 JAIN 36 JAISWAL 37 JASODI 38 JHHIMMER 39 JULAHA 40 KACHHI 41 KAHAR 42 KAHI 43 KALAR 44 KALI 45 KALRA 46 KANOJIA 47 KATNATAM 48 KEWAMKAT 49 KEWET 50 KOL 51 KSHTRIYA 52 KUMBHI 53 KUMHAR (POTTER) 54 KUMRAWAT 55 KUNVAL 56 KURMA 57 KURMI 58 KUSHWAHA 59 LODHI 60 LULAR 61 MAJHE -

Displacement and Development: Long Term Impacts
Displacement and Development: Long Term Impacts of the Partition of India ⇤ Prashant Bharadwaj† Rinchan Ali Mirza‡ Preliminary Draft as of September 2016. Please do not circulate. Abstract The partition of British India in 1947 resulted in one of the largest and most rapid migrations in human history. This paper examines how areas a↵ected by the partition fare in the long run. Using migrant presence as a proxy for the inten- sity of the impact of the partition, and district level data on agricultural output between 1911-2009, we find that areas that received more migrants have higher average yields, are more likely to take up high yielding varieties (HYV) of seeds, and are more likely to use agricultural technologies. These correlations are more pronounced after the Green Revolution in India. Using pre-partition data, we show that migrant placement is uncorrelated with soil conditions, agricultural infras- tructure, and agricultural yields prior to 1947; hence, the e↵ects are not solely explained by selective migration into districts with a higher potential for agricul- tural development. Migrants moving to India were more educated than both the natives who stayed and the migrants who moved out. Given the positive associa- tion of education with the adoption of high yielding varieties of seeds we highlight the presence of educated migrants during the timing of the Green Revolution as a potential pathway for the observed e↵ects. ⇤This project has benefitted from comments by Latika Chaudhary, James Fenske, Bishnupriya Gupta, Takashi Kurosaki, and participants at the World Economic History Congress, 2015. †University of California San Diego & NBER. -

Abdul En La India Ad Dharmi En La India Adaviar En La India Adi
Ore por los No-Alcanzados Ore por los No-Alcanzados Abdul en la India Ad Dharmi en la India País: India País: India Etnia: Abdul Etnia: Ad Dharmi Población: 35,000 Población: 1,035,000 Población Mundial: 63,000 Población Mundial: 1,035,000 Idioma Principal: Urdu Idioma Principal: Punjabi, Eastern Religión Principal: Islam Religión Principal: Hinduismo Estatus: Menos Alcanzado Estatus: Menos Alcanzado Seguidores de Cristo: Pocos, menos del 2% Seguidores de Cristo: Pocos, menos del 2% Biblia: Biblia Biblia: Biblia www.joshuaproject.net www.joshuaproject.net "Proclamen su gloria entre las naciones" Salmos 96:3 "Proclamen su gloria entre las naciones" Salmos 96:3 Ore por los No-Alcanzados Ore por los No-Alcanzados Adaviar en la India Adi Andhra en la India País: India País: India Etnia: Adaviar Etnia: Adi Andhra Población: 200 Población: 306,000 Población Mundial: 200 Población Mundial: 307,000 Idioma Principal: Tamil Idioma Principal: Telugu Religión Principal: Hinduismo Religión Principal: Hinduismo Estatus: Menos Alcanzado Estatus: Menos Alcanzado Seguidores de Cristo: Pocos, menos del 2% Seguidores de Cristo: Pocos, menos del 2% Biblia: Biblia Biblia: Biblia www.joshuaproject.net www.joshuaproject.net "Proclamen su gloria entre las naciones" Salmos 96:3 "Proclamen su gloria entre las naciones" Salmos 96:3 Ore por los No-Alcanzados Ore por los No-Alcanzados Adi Dravida en la India Adi Karnataka en la India País: India País: India Etnia: Adi Dravida Etnia: Adi Karnataka Población: 8,551,000 Población: 2,953,000 Población Mundial: 8,551,000 -

Partition and Postmemory in the Work of Kriti Arora and Sharlene Bamboat Rajee Paña Jejisher Gill a Thesis in the Department O
Partition and Postmemory in the Work of Kriti Arora and Sharlene Bamboat Rajee Paña Jejisher Gill A Thesis in The Department of Art History Presented in Partial Fulfillment of the Requirements for the Degree of Master of Arts (Art History) at Concordia University Montreal, Quebec, Canada April, 2014 © Rajee Paña Jejisher Gill, 2014 CONCORDIA UNIVERSITY School of Graduate Studies This is to certify that the thesis prepared By: Rajee Paña Jejisher Gill Entitled: Partition and Postmemory in the Work of Kriti Arora and Sharlene Bamboat and submitted in partial fulfillment of the requirements for the degree of Master of Arts (Art History) complies with the regulations of the University and meets the accepted standards with respect to originality and quality. Signed by the final Examining Committee: ____________________________________ Chair ____________________________________ Examiner Dr. Cynthia Hammond ____________________________________ Examiner Dr. Martha Langford ____________________________________ Examiner Dr. Alice Ming Wai Jim Approved By _____________________________ Dr. Anne Whitelaw Graduate Program Director ___________2014 _____________________________ Catherine Wild Dean of Faculty of Fine Arts iii Abstract Partition and Postmemory in the Work of Kriti Arora and Sharlene Bamboat Rajee Paña Jejisher Gill This thesis examines personal memory and inter- and transgenerational transmission, employing postmemory as a key interpretative lens to analyze the media work of New Delhi-based artist Kriti Arora’s five-minute silent film -

Patiala District, Punjab
GLOS3AHY OF CAS'rE NAIVllid RE'fURNED AT TH~ CEI'4SUS O}t' 1951 IN THE DISrRICTS OF PEPSU -_FOREWORD----_ .... ---- ..... __.... At the Oensus of 1951 there was a limited enumeration and tabulation of castes. Under the limited enumeration caste was recorded as returned by the respondent. Several complications arose out of this procedure. Many persons who returned their caste by generic or synonymous Scheduled Caste or tribe names not found in the prescribed lists were left out of the count of Scheduled Castes and Tribes and are-count had to be later ordered in some states. The Backward Classes Commission could not be provided with the 1951 population of individual castes and tribes or their individua~ educational and economic characteristics. 2. If a complete enumeration of castes is ordered at the next census much preliminary study will have to be carried out in order to ensure correct and rational enumeration and tabulation. For that purpose and, in fact, for any systematic enumeration of castes, a glossary of caste names as returned at the 1951 Census would be invaluable. This explains the prepara~ion of t~is glossary. 3. The Glossary has been prepared by running through all the male slips relating to the non-backward classes and the five per cent sample male slips of Backward Classes and sh~uld, therefore, be a complete list of caste names. All caste names as found in the slips have been deliberately included in the Glossary without any attempt at rationalisation. Many of these names are synonymous; some relate to sub-castes or gotras; and some are only generic names.