Protein Structure
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Side-Chain Liquid Crystal Polymers (SCLCP): Methods and Materials. an Overview
Materials 2009, 2, 95-128; doi:10.3390/ma2010095 OPEN ACCESS materials ISSN 1996-1944 www.mdpi.com/journal/materials Review Side-chain Liquid Crystal Polymers (SCLCP): Methods and Materials. An Overview Tomasz Ganicz * and Włodzimierz Stańczyk Centre of Molecular and Macromolecular Studies, Polish Academy of Science, Sienkiewicza 112, 90- 363 Łódź, Poland; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel. +48-42-684-71-13; Fax: +48-42-684-71-26 Received: 5 February 2009; in revised form: 3 March 2009 / Accepted: 9 March 2009 / Published: 11 March 2009 Abstract: This review focuses on recent developments in the chemistry of side chain liquid crystal polymers. It concentrates on current trends in synthetic methods and novel, well defined structures, supramolecular arrangements, properties, and applications. The review covers literature published in this century, apart from some areas, such as dendritic and elastomeric systems, which have been recently reviewed. Keywords: Liquid crystals, side chain polymers, polymer modification, polymer synthesis, self-assembly. 1. Introduction Over thirty years ago the work of Finkelmann and Ringsdorf [1,2] gave important momentum to synthesis of side chain liquid crystal polymers (SCLCPs), materials which combine the anisotropy of liquid crystalline mesogens with the mechanical properties of polymers. Although there were some earlier attempts [2], their approach of decoupling the motions of a polymer main chain from a mesogen, thus allowed side chain moieties to build up long range ordering (Figure 1). They were able to synthesize polymers with nematic, smectic and cholesteric phases via free radical polymerization of methacryloyl type monomers [3]. -

Glossary - Cellbiology
1 Glossary - Cellbiology Blotting: (Blot Analysis) Widely used biochemical technique for detecting the presence of specific macromolecules (proteins, mRNAs, or DNA sequences) in a mixture. A sample first is separated on an agarose or polyacrylamide gel usually under denaturing conditions; the separated components are transferred (blotting) to a nitrocellulose sheet, which is exposed to a radiolabeled molecule that specifically binds to the macromolecule of interest, and then subjected to autoradiography. Northern B.: mRNAs are detected with a complementary DNA; Southern B.: DNA restriction fragments are detected with complementary nucleotide sequences; Western B.: Proteins are detected by specific antibodies. Cell: The fundamental unit of living organisms. Cells are bounded by a lipid-containing plasma membrane, containing the central nucleus, and the cytoplasm. Cells are generally capable of independent reproduction. More complex cells like Eukaryotes have various compartments (organelles) where special tasks essential for the survival of the cell take place. Cytoplasm: Viscous contents of a cell that are contained within the plasma membrane but, in eukaryotic cells, outside the nucleus. The part of the cytoplasm not contained in any organelle is called the Cytosol. Cytoskeleton: (Gk. ) Three dimensional network of fibrous elements, allowing precisely regulated movements of cell parts, transport organelles, and help to maintain a cell’s shape. • Actin filament: (Microfilaments) Ubiquitous eukaryotic cytoskeletal proteins (one end is attached to the cell-cortex) of two “twisted“ actin monomers; are important in the structural support and movement of cells. Each actin filament (F-actin) consists of two strands of globular subunits (G-Actin) wrapped around each other to form a polarized unit (high ionic cytoplasm lead to the formation of AF, whereas low ion-concentration disassembles AF). -

Heterogeneous Impacts of Protein-Stabilizing Osmolytes On
bioRxiv preprint doi: https://doi.org/10.1101/328922; this version posted May 23, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Heterogeneous Impacts of Protein-Stabilizing Osmolytes on Hydrophobic Interaction Mrinmoy Mukherjee and Jagannath Mondal⇤ Tata Institute of Fundamental Research Hyderabad, 500107 India E-mail: [email protected],+914020203091 Abstract Osmolytes’ mechanism of protecting proteins against denaturation is a longstanding puzzle, further complicated by the complex diversities inherent in protein sequences. An emergent approach in understanding osmolytes’ mechanism of action towards biopoly- mer has been to investigate osmolytes’ interplay with hydrophobic interaction, the ma- jor driving force of protein folding. However, the crucial question is whether all these protein-stabilizing osmolytes display a single unified mechanism towards hydrophobic interactions. By simulating the hydrophobic collapse of a macromolecule in aqueous solutions of two such osmoprotectants, Glycine and Trimethyl N-oxide (TMAO), both of which are known to stabilize protein’s folded conformation, we here demonstrate that these two osmolytes can impart mutually contrasting effects towards hydropho- bic interaction. While TMAO preserves its protectant nature across diverse range of polymer-osmolyte interactions, glycine is found to display an interesting cross-over from being a protectant at weaker polymer-osmolyte interaction to a denaturant of hydrophobicity at stronger polymer-osmolyte interactions. A preferential-interaction analysis reveals that a subtle balance of conformation-dependent exclusion/binding of ⇤To whom correspondence should be addressed 1 bioRxiv preprint doi: https://doi.org/10.1101/328922; this version posted May 23, 2018. -

The Role of Side-Chain Branch Position on Thermal Property of Poly-3-Alkylthiophenes
Polymer Chemistry The Role of Side-chain Branch Position on Thermal Property of Poly-3-alkylthiophenes Journal: Polymer Chemistry Manuscript ID PY-ART-07-2019-001026.R2 Article Type: Paper Date Submitted by the 02-Oct-2019 Author: Complete List of Authors: Gu, Xiaodan; University of Southern Mississippi, School of Polymer Science and Engineering Cao, Zhiqiang; University of Southern Mississippi, School of Polymer Science and Engineering Galuska, Luke; University of Southern Mississippi, School of Polymer Science and Engineering Qian, Zhiyuan; University of Southern Mississippi, School of Polymer Science and Engineering Zhang, Song; University of Southern Mississippi, School of Polymer Science and Engineering Huang, Lifeng; University of Southern Mississippi, School of Polymers and High Performance Materials Prine, Nathaniel; University of Southern Mississippi, School of Polymer Science and Engineering Li, Tianyu; Oak Ridge National Laboratory; University of Tennessee Knoxville He, Youjun; Oak Ridge National Laboratory Hong, Kunlun; Oak Ridge National Laboratory, Center for Nanophase Materials Science; Oak Ridge National Laboratory, Center for Nanophase Materials Science Page 1 of 13 PleasePolymer do not Chemistryadjust margins ARTICLE The Role of Side-chain Branch Position on Thermal Property of Poly-3-alkylthiophenes Received 00th January 20xx, a a a a a a Accepted 00th January 20xx Zhiqiang Cao, Luke Galuska, Zhiyuan Qian, Song Zhang, Lifeng Huang, Nathaniel Prine, Tianyu Li,b, c Youjun He,b Kunlun Hong,*b, c and Xiaodan Gu*a DOI: 10.1039/x0xx00000x Thermomechanical properties of conjugated polymers (CPs) are greatly influenced by both their microstructures and backbone structures. In the present work, to investigate the effect of side-chain branch position on the backbone’s mobility and molecular packing structure, four poly (3-alkylthiophene-2, 5-diyl) derivatives (P3ATs) with different side chains, either branched and linear, were synthesized by a quasi-living Kumada catalyst transfer polymerization (KCTP) method. -

Protein Structure & Folding
6 Protein Structure & Folding To understand protein folding In the last chapter we learned that proteins are composed of amino acids Goal as a chemical equilibrium. linked together by peptide bonds. We also learned that the twenty amino acids display a wide range of chemical properties. In this chapter we will see Objectives that how a protein folds is determined by its amino acid sequence and that the three-dimensional shape of a folded protein determines its function by After this chapter, you should be able to: the way it positions these amino acids. Finally, we will see that proteins fold • describe the four levels of protein because doing so minimizes Gibbs free energy and that this minimization structure and the thermodynamic involves both making the most favorable bonds and maximizing disorder. forces that stabilize them. • explain how entropy (S) and enthalpy Proteins exhibit four levels of structure (H) contribute to Gibbs free energy. • use the equation ΔG = ΔH – TΔS The structure of proteins can be broken down into four levels of to determine the dependence of organization. The first is primary structure, the linear sequence of amino the favorability of a reaction on acids in the polypeptide chain. By convention, the primary sequence is temperature. written in order from the amino acid at the N-terminus (by convention • explain the hydrophobic effect and its usually on the left) to the amino acid at the C-terminus. The second level role in protein folding. of protein structure, secondary structure, is the local conformation adopted by stretches of contiguous amino acids. -

DNA Glycosylase Exercise - Levels 1 & 2: Answer Key
Name________________________ StarBiochem DNA Glycosylase Exercise - Levels 1 & 2: Answer Key Background In this exercise, you will explore the structure of a DNA repair protein found in most species, including bacteria. DNA repair proteins move along DNA strands, checking for mistakes or damage. DNA glycosylases, a specific type of DNA repair protein, recognize DNA bases that have been chemically altered and remove them, leaving a site in the DNA without a base. Other proteins then come along to fill in the missing DNA base. Learning objectives We will explore the relationship between a protein’s structure and its function in a human DNA glycosylase called human 8-oxoguanine glycosylase (hOGG1). Getting started We will begin this exercise by exploring the structure of hOGG1 using a molecular 3-D viewer called StarBiochem. In this particular structure, the repair protein is bound to a segment of DNA that has been damaged. We will first focus on the structure hOGG1 and then on how this protein interacts with DNA to repair a damaged DNA base. • To begin using StarBiochem, please navigate to: http://mit.edu/star/biochem/. • Click on the Start button Click on the Start button for StarBiochem. • Click Trust when a prompt appears asking if you trust the certificate. • In the top menu, click on Samples à Select from Samples. Within the Amino Acid/Proteins à Protein tab, select “DNA glycosylase hOGG1 w/ DNA – H. sapiens (1EBM)”. “1EBM” is the four character unique ID for this structure. Take a moment to look at the structure from various angles by rotating and zooming on the structure. -
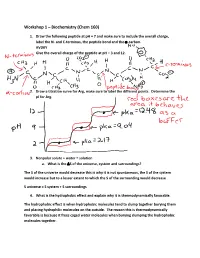
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

Improved Prediction of Protein Side-Chain Conformations with SCWRL4 Georgii G
proteins STRUCTURE O FUNCTION O BIOINFORMATICS Improved prediction of protein side-chain conformations with SCWRL4 Georgii G. Krivov,1,2 Maxim V. Shapovalov,1 and Roland L. Dunbrack Jr.1* 1 Institute for Cancer Research, Fox Chase Cancer Center, 333 Cottman Avenue, Philadelphia, Pennsylvania 19111 2 Department of Applied Mathematics, Moscow Engineering Physics Institute (MEPhI), Kashirskoe Shosse 31, Moscow 115409, Russian Federation INTRODUCTION ABSTRACT The side-chain conformation prediction problem is an integral Determination of side-chain conformations is an component of protein structure determination, protein structure important step in protein structure prediction and protein design. Many such methods have prediction, and protein design. In single-site mutants and in closely been presented, although only a small number related proteins, the backbone often changes little and structure pre- 1 are in widespread use. SCWRL is one such diction can be accomplished by accurate side-chain prediction. In method, and the SCWRL3 program (2003) has docking of ligands and other proteins, taking into account changes remained popular because of its speed, accuracy, in side-chain conformation is often critical to accurate structure pre- and ease-of-use for the purpose of homology dictions of complexes.2–4 Even in methods that take account of modeling. However, higher accuracy at compara- changes in backbone conformation, one step in the process is recal- ble speed is desirable. This has been achieved in culation of side-chain conformation or ‘‘repacking.’’5 Because many a new program SCWRL4 through: (1) a new backbone conformations may be sampled in model refinements, backbone-dependent rotamer library based on side-chain prediction must also be very fast. -

Unit 2 Cell Biology Page 1 Sub-Topics Include
Sub-Topics Include: 2.1 Cell structure 2.2 Transport across cell membranes 2.3 Producing new cells 2.4 DNA and the production of proteins 2.5 Proteins and enzymes 2.6 Genetic Engineering 2.7 Respiration 2.8 Photosynthesis Unit 2 Cell Biology Page 1 UNIT 2 TOPIC 1 1. Cell Structure 1. Types of organism Animals and plants are made up of many cells and so are described as multicellular organisms. Similar cells doing the same job are grouped together to form tissues. Animals and plants have systems made up of a number of organs, each doing a particular job. For example, the circulatory system is made up of the heart, blood and vessels such as arteries, veins and capillaries. Some fungi such as yeast are single celled while others such as mushrooms and moulds are larger. Cells of animals plants and fungi have contain a number of specialised structures inside their cells. These specialised structures are called organelles. Bacteria are unicellular (exist as single cells) and do not have organelles in the same way as other cells. 2. Looking at Organelles Some organelles can be observed by staining cells and viewing them using a light microscope. Other organelles are so small that they cannot be seen using a light microscope. Instead, they are viewed using an electron microscope, which provides a far higher magnification than a light microscope. Most organelles inside cells are surrounded by a membrane which is similar in composition (make-up) to the cell membrane. Unit 2 Cell Biology Page 2 3. A Typical Cell ribosome 4. -

Collagen and Creatine
COLLAGEN AND CREATINE : PROTEIN AND NONPROTEIN NITROGENOUS COMPOUNDS Color index: . Important . Extra explanation “ THERE IS NO ELEVATOR TO SUCCESS. YOU HAVE TO TAKE THE STAIRS ” 435 Biochemistry Team • Amino acid structure. • Proteins. • Level of protein structure. RECALL: 435 Biochemistry Team Amino acid structure 1- hydrogen atom *H* ( which is distictive for each amino 2- side chain *R* acid and gives the amino acid a unique set of characteristic ) - Carboxylic acid group *COOH* 3- two functional groups - Primary amino acid group *NH2* ( except for proline which has a secondary amino acid) .The amino acid with a free amino Group at the end called “N-Terminus” . Alpha carbon that is attached to: to: thatattachedAlpha carbon is .The amino acid with a free carboxylic group At the end called “ C-Terminus” Proteins Proteins structure : - Building blocks , made of small molecules unit called amino acid which attached together in long chain by a peptide bond . Level of protein structure Tertiary Quaternary Primary secondary Single amino acids Region stabilized by Three–dimensional attached by hydrogen bond between Association of covalent bonds atoms of the polypeptide (3D) shape of called peptide backbone. entire polypeptide multi polypeptides chain including forming a bonds to form a Examples : linear sequence of side chain (R functional protein. amino acids. Alpha helix group ) Beta sheet 435 Biochemistry Team Level of protein structure 435 Biochemistry Team Secondary structure Alpha helix: - It is right-handed spiral , which side chain extend outward. - it is stabilized by hydrogen bond , which is formed between the peptide bond carbonyl oxygen and amide hydrogen. - each turn contains 3.6 amino acids. -

Amino Acid Building Block Models – in Brief
Amino Acid Building Block Models – In Brief Key Teaching Points for Amino Acid Building Block Models© Overall Student Learning Objective: What Dictates How a Protein Folds? Amino acids are the building blocks of proteins. All amino acids have an identical core structure consisting of an alpha-carbon, carboxyl group, amino group and R-group (sidechain). A linear chain of amino acids is a polypeptide. The primary sequence of a protein is the linear sequence of amino acids in a polypeptide. Proteins are made up of amino acid monomers linked together by peptide bonds. Peptide bond formation between amino acids results in the release of water (dehydration synthesis or condensation reaction). The protein backbone is characterized by the “N-C-C-N-C-C. .” pattern. The “ends” of the protein can be identified by the N-terminus (amino group) end and the C-terminus (carboxyl group) end. For a more complete lesson guide, please visit: http://www.3dmoleculardesigns.com/3DMD-Files/AABB/ContentsandAssembly.pdf Amino Acid Core Structure Build an amino acid according to the diagram to the right: 1. Identify the alpha carbon, amino group, carboxyl group and R-group (sidechain representation) in the structure you have constructed. Two amino acids can be chemically linked by a reaction called “condensation” or “dehydration synthesis” to form a dipeptide bond linking the two amino acids. A chain of amino acid units (monomers) linked together by peptide bonds is called a polypeptide. General Dipeptide Structure Construct a model of a dipeptide using the amino acid models previously built. 2. What are the products of the condensation reaction (dehydration synthesis)? 3. -

Proteasomes on the Chromosome Cell Research (2017) 27:602-603
602 Cell Research (2017) 27:602-603. © 2017 IBCB, SIBS, CAS All rights reserved 1001-0602/17 $ 32.00 RESEARCH HIGHLIGHT www.nature.com/cr Proteasomes on the chromosome Cell Research (2017) 27:602-603. doi:10.1038/cr.2017.28; published online 7 March 2017 Targeted proteolysis plays an this process, both in mediating homolog ligases, respectively, as RN components important role in the execution and association and in providing crossovers that impact crossover formation [5, 6]. regulation of many cellular events. that tether homologs and ensure their In the first of the two papers, Ahuja et Two recent papers in Science identify accurate segregation. Meiotic recom- al. [7] report that budding yeast pre9∆ novel roles for proteasome-mediated bination is initiated by DNA double- mutants, which lack a nonessential proteolysis in homologous chromo- strand breaks (DSBs) at many sites proteasome subunit, display defects in some pairing, recombination, and along chromosomes, and the multiple meiotic DSB repair, in chromosome segregation during meiosis. interhomolog interactions formed by pairing and synapsis, and in crossover Protein degradation by the 26S pro- DSB repair drive homolog association, formation. Similar defects are seen, teasome drives a variety of processes culminating in the end-to-end homolog but to a lesser extent, in cells treated central to the cell cycle, growth, and dif- synapsis by a protein structure called with the proteasome inhibitor MG132. ferentiation. Proteins are targeted to the the synaptonemal complex (SC) [3]. These defects all can be ascribed to proteasome by covalently ligated chains SC-associated focal protein complexes, a failure to remove nonhomologous of ubiquitin, a small (8.5 kDa) protein.