Can Quantifying Free-Circulating DNA in Plasma Be Used to Identify Subjects with High-Grade Pre-Invasive Endobronchial Lesions?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Markers in Lung Cancer Diagnosis: a Review
International Journal of Molecular Sciences Review Genetic Markers in Lung Cancer Diagnosis: A Review Katarzyna Wadowska 1 , Iwona Bil-Lula 1 , Łukasz Trembecki 2,3 and Mariola Sliwi´ ´nska-Mosso´n 1,* 1 Department of Medical Laboratory Diagnostics, Division of Clinical Chemistry and Laboratory Haematology, Wroclaw Medical University, 50-556 Wroclaw, Poland; [email protected] (K.W.); [email protected] (I.B.-L.) 2 Department of Radiation Oncology, Lower Silesian Oncology Center, 53-413 Wroclaw, Poland; [email protected] 3 Department of Oncology, Faculty of Medicine, Wroclaw Medical University, 53-413 Wroclaw, Poland * Correspondence: [email protected]; Tel.: +48-71-784-06-30 Received: 1 June 2020; Accepted: 25 June 2020; Published: 27 June 2020 Abstract: Lung cancer is the most often diagnosed cancer in the world and the most frequent cause of cancer death. The prognosis for lung cancer is relatively poor and 75% of patients are diagnosed at its advanced stage. The currently used diagnostic tools are not sensitive enough and do not enable diagnosis at the early stage of the disease. Therefore, searching for new methods of early and accurate diagnosis of lung cancer is crucial for its effective treatment. Lung cancer is the result of multistage carcinogenesis with gradually increasing genetic and epigenetic changes. Screening for the characteristic genetic markers could enable the diagnosis of lung cancer at its early stage. The aim of this review was the summarization of both the preclinical and clinical approaches in the genetic diagnostics of lung cancer. The advancement of molecular strategies and analytic platforms makes it possible to analyze the genome changes leading to cancer development—i.e., the potential biomarkers of lung cancer. -

Invasive Cervical Cancer Audit; EU Guidelines for Quality Assurance
The 4th EFCS Annual Tutorial Ospedale Universitario di Cattinara, Strada di Fiume, Trieste Handouts for lectures and workshops – I I - Gynaecological cytopathology Mrs Rietje Salet‐van‐de Pol, Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands Gynecological cytology: technical aspects ............................................................................... 2 Non‐neoplastic gynecological cytology .................................................................................... 6 • Dr Giovanni Negri, General Hospital of Bolzano, Bozano SIL and cancer; ASC‐US, ASC‐H, diagnostic pitfalls and look‐alikes; glandular abnormalities 11 • Dr Amanda Herbert, Guy’s & St Thomas’ NHS Foundation Trust, London Invasive cervical cancer audit; EU guidelines for quality assurance ...................................... 17 1 Gynecological cytology: technical aspects Rietje Salet-van de Pol Important in specimen processing is to obtain as much as possible well preserved cells for microscopically evaluation. The quality of the smear depends on cell sampling, fixation and staining. For obtaining enough cervical material you are dependent on the cell sampler. For cervical cytology two types of specimen are available: conventional smears and liquid based cytology (LBC). Conventional, Thinprep and Surepath slides In conventional cytology the cell sampler makes the smear and is responsible for the fixation of the cells. Reasons for unsatisfactory conventional smears can be obscuring blood or inflammatory cells, thick smears with overlapping cells, poor preservation of the cells due to late fixation and low cellularity. In LBC the cell sampler immediately transferred the cellular material into a vial with fixative (fixating solution) which gives a better preservation of the cells. The laboratory is responsible for processing of the smear. LBC gives equally distribution of the cells in a thin cell layer of well preserved cells. The rate of unsatisfactory smears is lower. -
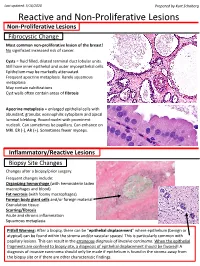
Reactive and Non-Proliferative Lesions
Last updated: 5/16/2020 Prepared by Kurt Schaberg Reactive and Non-Proliferative Lesions Non-Proliferative Lesions Fibrocystic Change Most common non-proliferative lesion of the breast! No significant increased risk of cancer. Cysts = fluid filled, dilated terminal duct lobular units. Still have inner epithelial and outer myoepithelial cells. Epithelium may be markedly attenuated. Frequent apocrine metaplasia. Rarely squamous metaplasia May contain calcifications Cyst walls often contain areas of fibrosis Apocrine metaplasia = enlarged epithelial cells with abundant, granular, eosinophilic cytoplasm and apical luminal blebbing. Round nuclei with prominent nucleoli. Can sometimes be papillary. Can enhance on MRI. ER (-), AR (+). Sometimes fewer myoeps. Inflammatory/Reactive Lesions Biopsy Site Changes Changes after a biopsy/prior surgery. Frequent changes include: Organizing hemorrhage (with hemosiderin laden macrophages and blood) Fat necrosis (with foamy macrophages) Foreign body giant cells and/or foreign material Granulation tissue Scarring/fibrosis Acute and chronic inflammation Squamous metaplasia Pitfall Warning: After a biopsy, there can be “epithelial displacement” where epithelium (benign or atypical) can be found within the stroma and/or vascular spaces! This is particularly common with papillary lesions. This can result in the erroneous diagnosis of invasive carcinoma. When the epithelial fragments are confined to biopsy site, a diagnosis of epithelial displacement should be favored! A diagnosis of invasive carcinoma should -

Primary Immature Teratoma of the Thigh Fig
CORRESPONDENCE 755 8. Gray W, Kocjan G. Diagnostic Cytopathology. 2nd ed. London: Delete all that do not apply: Elsevier Health Sciences, 2003; 677. 9. Richards A, Dalrymple C. Abnormal cervicovaginal cytology, unsatis- Cervix, colposcopic biopsy/LLETZ/cone biopsy: factory colposcopy and the use of vaginal estrogen cream: an obser- vational study of clinical outcomes for women in low estrogen states. Diagnosis: NIL (No intraepithelial lesion WHO 2014) J Obstet Gynaecol Res 2015; 41: 440e4. LSIL (CIN 1 with HPV effect WHO 2014) 10. Darragh TM, Colgan TJ, Cox T, et al. The lower anogenital squamous HSIL (CIN2/3 WHO 2014) terminology standardization project for HPV-associated lesions: back- Squamous cell carcinoma ground and consensus recommendation from the College of American Immature squamous metaplasia Pathologists and the American Society for Colposcopy and Cervical Adenocarcinoma in situ (AIS, HGGA) e Adenocarcinoma Pathology. Arch Pathol Lab Med 2012; 136: 1267 97. Atrophic change 11. McCluggage WG. Endocervical glandular lesions: controversial aspects e Extending into crypts: Not / Idenfied and ancillary techniques. J Clin Pathol 2013; 56: 164 73. Epithelial stripping: Not / Present 12. World Health Organization (WHO). Comprehensive Cervical Cancer Invasive disease: Not / Idenfied / Micro-invasive Control: A Guide to Essential Practice. 2nd ed. Geneva: WHO, 2014. Depth of invasion: mm Transformaon zone: Not / Represented Margins: DOI: https://doi.org/10.1016/j.pathol.2019.07.014 Ectocervical: Not / Clear Endocervical: Not / Clear Circumferenal: Not / Clear p16 status: Negave / Posive Primary immature teratoma of the thigh Fig. 3 A proposed synoptic reporting format for pathologists reporting colposcopic biopsies and cone biopsies or LLETZ. Sir, Teratomas are germ cell tumours composed of a variety of HSIL, AIS, micro-invasive or more advanced invasive dis- somatic tissues derived from more than one germ layer 12 ease. -

Squamous Metaplasia of the Tracheal Epithelium in Children
Thorax: first published as 10.1136/thx.31.2.167 on 1 April 1976. Downloaded from Thorax (1976), 31, 167. Squamous metaplasia of the tracheal epithelium in children AVINASH MITHAL' and JOHN L. EMERY2 The Chest Clinic, Lincoln' and The Children's Hospital, Sheffield' Mithal, A. and Emery, J. L. (1976). Thorax, 31, 167-171. Squamous metaplasia of the tracheal epithelium in children. Thirty-seven (16%) tracheas from 2170 children showed squamous metaplasia. (Cases with tracheo-oesophageal fistula and congenital heart disease were excluded.) The metaplasia extended into the bronchi in 15 cases. Features of pulmonary retention were present in seven cases. Respiratory infection, probably viral, seemed to be the most significant causative factor in 20 children, including those with cystic fibrosis. Tracheal instrumentation was a possible factor in 11 cases but oxygen therapy alone did not seem important. The metaplasia was almost certainly congenital in one child and probably in two others but no stillborn infants showed metaplasia. In many children the metaplasia seemed to be due to a combination of factors. Squamous metaplasia of the trachea in childhood Tracheas from children with tracheo-oesophageal has been described in cases of measles (Gold- fistula and those with congenital heart disease or zieher, 1918), influenza (Askanazy, 1919), cystic other gross deformities were excluded. There were fibrosis of the pancreas (Zuelzer and Newton, thus 2331 tracheas available for study. Epithelium 1949), and following intubation of the trachea was absent in 16 cases. This left 2170 tracheas for http://thorax.bmj.com/ (Rasche and Kuhns, 1972) and tracheostomy histological analysis. (Sara, 1967; Sara and Reye, 1969). -

Squamous Metaplasia of Normal and Carcinoma in Situ of HPV 16-Immortalized Human Endocervical Cells1
[CANCER RESEARCH 52. 4254-4260, August I, 1992] Squamous Metaplasia of Normal and Carcinoma in Situ of HPV 16-Immortalized Human Endocervical Cells1 Qi Sun, Kouichiro Tsutsumi, M. Brian Kelleher, Alan Pater, and Mary M. Pater2 Division of Basic Medical Sciences, Faculty of Medicine, Memorial University of Newfoundland, St. John's, Newfoundland, Canada A1B ÌV6 ABSTRACT genomic DNA, most frequently of HPV 16, has been detected in 90% of the cervical carcinomas and are found to be actively The importance of cervical squamous metaplasia and human papil- expressed (6, 7). HPV 16 DNA has been used to transform lomavirus 16 (HPV 16) infection for cervical carcinoma has been well human foreskin and ectocervical keratinocytes (8, 9). It immor established. Nearly 87% of the intraepithelial neoplasia of the cervix occur in the transformation zone, which is composed of squamous meta- talizes human keratinocytes efficiently, producing cell clones plastic cells with unclear origin. HPV DNA, mostly HPV 16, has been with indefinite life span in culture. Different approaches have found in 90% of cervical carcinomas, but only limited experimental data been taken to examine the behavior of these immortalized cell are available to discern the role of HPV 16 in this tissue specific onco- lines in conditions allowing squamous differentiation (10, 11). genesis. We have initiated in vivo studies of cultured endocervical cells After transplantation in vivo, the HPV 16-immortalized kerat as an experimental model system for development of cervical neoplasia. inocytes retain thépotential for squamous differentiation, Using a modified in vivo implantation system, cultured normal endocer forming abnormal epithelium without dysplastic changes at vical epithelial cells formed epithelium resembling squamous metapla early passages and with various dysplastic changes only after sia, whereas those immortalized by HPV 16 developed into lesions long periods of time in culture (10). -

Squamous Cell Carcinoma of the Breast As a Clinical Diagnostic Challenge
582 MOLECULAR AND CLINICAL ONCOLOGY 8: 582-586, 2018 Squamous cell carcinoma of the breast as a clinical diagnostic challenge KATARZYNA JAKUBOWSKA1, LUIZA KAŃCZUGA‑KODA1, WOJCIECH KISIELEWSKI2, MARIUSZ KODA3 and WALDEMAR FAMULSKI1,2 1Department of Pathomorphology, Comprehensive Cancer Center, 15‑027 Białystok; Departments of 2Medical Pathomorphology and 3General Pathomorphology, Medical University of Białystok, 15‑269 Białystok, Poland Received September 17, 2017; Accepted December 14, 2017 DOI: 10.3892/mco.2018.1581 Abstract. Squamous cell carcinoma (SqCC) of the breast metaplasia of ductal and lobular epithelial cells can be should be differentiated between the primary skin keratinizing linked with fat necrosis and infracted ademonas. Squamous squamous carcinoma and squamous metaplastic cancer. In the cell carcinoma should be differentiated between lesions of current study, the cases of two patients who were diagnosed keratinizing squamous carcinoma and squamous metaplasia with SqCC originated from skin and the breast were discussed. associated to mammary carcinoma (2). The characteristic A fine-needle aspiration biopsy confirmed the presence features of metaplastic cell carcinoma include: i) primary of atypical squamous cells. In both cases, the microscopic carcinoma without other neoplastic components such as ductal examination of the surgical specimen revealed a malignant or mesenchymal elements, ii) the tumor origin is independent neoplasm differentiated into SqCC characterized by keratin- of the overlying skin and nipple and iii) absence of primary izing cancer cells with abundant eosiphilic cytoplasm with epidermoid tumors present in other site (oral cavity, bronchus, large, hyperchromatic vesicular nuclei. Immunohistochemical esophagus, bladder, cervix ect.) (3). However, squamous studies showed negative for progesterone and estrogen recep- metaplastic carcinoma should be also differentiated with pure tors and human epidermal growth factor receptor 2. -

Chapter 1 Cellular Reaction to Injury 3
Schneider_CH01-001-016.qxd 5/1/08 10:52 AM Page 1 chapter Cellular Reaction 1 to Injury I. ADAPTATION TO ENVIRONMENTAL STRESS A. Hypertrophy 1. Hypertrophy is an increase in the size of an organ or tissue due to an increase in the size of cells. 2. Other characteristics include an increase in protein synthesis and an increase in the size or number of intracellular organelles. 3. A cellular adaptation to increased workload results in hypertrophy, as exemplified by the increase in skeletal muscle mass associated with exercise and the enlargement of the left ventricle in hypertensive heart disease. B. Hyperplasia 1. Hyperplasia is an increase in the size of an organ or tissue caused by an increase in the number of cells. 2. It is exemplified by glandular proliferation in the breast during pregnancy. 3. In some cases, hyperplasia occurs together with hypertrophy. During pregnancy, uterine enlargement is caused by both hypertrophy and hyperplasia of the smooth muscle cells in the uterus. C. Aplasia 1. Aplasia is a failure of cell production. 2. During fetal development, aplasia results in agenesis, or absence of an organ due to failure of production. 3. Later in life, it can be caused by permanent loss of precursor cells in proliferative tissues, such as the bone marrow. D. Hypoplasia 1. Hypoplasia is a decrease in cell production that is less extreme than in aplasia. 2. It is seen in the partial lack of growth and maturation of gonadal structures in Turner syndrome and Klinefelter syndrome. E. Atrophy 1. Atrophy is a decrease in the size of an organ or tissue and results from a decrease in the mass of preexisting cells (Figure 1-1). -

Download Download
JOURNAL OF THE ITALIAN SOCIETY OF ANATOMIC PATHOLOGY AND DIAGNOSTIC CYTOPATHOLOGY, ITALIAN DIVISION OF THE INTERNATIONAL ACADEMY OF PATHOLOGY Periodico trimestrale - Aut. Trib. di Genova n. 75 del 22/06/1949 ISSN: 1591-951X (Online) The GIPAD handbook of the gastrointestinal pathologist (in the Covid-19 era) - Part I 03VOL. 112 Edited by Paola Parente and Matteo Fassan SEPTEMBER 2020 Editor-in-Chief C. Doglioni G. Pelosi M. Barbareschi San Raffaele Scientific Institute, Milan University of Milan Service of Anatomy and M. Fassan F. Pierconti University of Padua Pathological Histology, Trento Catholic University of Sacred G. Fornaciari Heart, Rome Associate Editor University of Pisa M. Chilosi M.P. Foschini S. Pileri Department of Pathology, Verona Bellaria Hospital, Bologna Milano European Institute of University, Verona G. Fraternali Orcioni Oncology, Milan S. Croce e Carle Hospital, Cuneo 03Vol. 112 P. Querzoli Managing Editor E. Fulcheri St Anna University Hospital, Ferrara University of Genoa September 2020 P. N oz za L. Resta M. Guido Pathology Unit, Ospedali Galliera, University of Bari Genova, Italy University of Padua S. Lazzi G. Rindi Catholic University of Sacred Italian Scientific Board University of Siena L. Leoncini M. Brunelli Heart, Rome University of Siena E.D. Rossi University of Verona C. Luchini G. Bulfamante Catholic University of Sacred University of Verona University of Milano G. Magro Heart, Rome G. Cenacchi University of Catania A.G. Rizzo University of Bologna E. Maiorano “Villa Sofia-Cervello” Hospital, C. Clemente University of Bari Aldo Moro Palermo San Donato Hospital, Milano A. Marchetti G. Rossi M. Colecchia University of Chieti-Pescara Hospital S. -

Current and Future Development in Lung Cancer Diagnosis
International Journal of Molecular Sciences Review Current and Future Development in Lung Cancer Diagnosis Reem Nooreldeen and Horacio Bach * Division of Infectious Diseases, Faculty of Medicine, The University of British Columbia, Vancouver, BC V6H 3Z6, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-604-875-4111 (ext. 62107) Abstract: Lung cancer is the leading cause of cancer-related deaths in North America and other developed countries. One of the reasons lung cancer is at the top of the list is that it is often not diagnosed until the cancer is at an advanced stage. Thus, the earliest diagnosis of lung cancer is crucial, especially in screening high-risk populations, such as smokers, exposure to fumes, oil fields, toxic occupational places, etc. Based on the current knowledge, it looks that there is an urgent need to identify novel biomarkers. The current diagnosis of lung cancer includes different types of imaging complemented with pathological assessment of biopsies, but these techniques can still not detect early lung cancer developments. In this review, we described the advantages and disadvantages of current methods used in diagnosing lung cancer, and we provide an analysis of the potential use of body fluids as carriers of biomarkers as predictors of cancer development and progression. Keywords: lung cancer; diagnosis; imaging; biomarkers; predictors; body fluids 1. Introduction Lung cancer is the most common cause of cancer-related deaths in North America Citation: Nooreldeen, R.; Bach, H. and other developed countries. According to the 2020 special report on lung cancer, this Current and Future Development in disease is the most commonly diagnosed cancer and the leading cause of cancer death in Lung Cancer Diagnosis. -

Tracheopathia Osteoplastica
J Clin Pathol: first published as 10.1136/jcp.20.6.814 on 1 November 1967. Downloaded from J. clin. Path. (1967), 20, 814 Tracheopathia osteoplastica S. P. B. WAY' From the Department ofPathology, St Mary's Hospital Medical School, London SYNOPSIS Only four cases of tracheopathia osteoplastica have been reported in Britain in the past 110 years. Ten further cases are now reported; in two the diagnosis was made at bronchoscopy. It is suggested that the reputed rarity of the condition in Britain is not the whole picture. Tracheopathia osteoplastica is a condition in which there is an accumulation of bony and cartilaginous nodules beneath the tracheal mucosa, sometimes spreading into the main bronchi, and occasionally affecting the larynx. More than 120 cases have been described in the world literature, including some distinguished accounts of the disease, but in Britain only three cases (Bowen, 1959; Salm, 1961; Baird and Macartney, 1966) have been reported since 1857 when Samuel Wilks, physician and morbid anato- copyright. mist to Guy's Hospital, first described ossific de- a posits on the larynx, trachea, and bronchi in a man aged 38 who died of phthisis. Ten further cases can now be reported. In 1952, a woman aged 61, who had collapsed and died at work, was brought to St Mary's Hospital, where the cause of death was found to be coronary occlusion; tracheopathia osteoplastica was present http://jcp.bmj.com/ and the specimen was preserved (case 1, Fig. 1). Ten years later, a man aged 51 died after a coronary thrombosis and again, at necropsy, tracheopathia osteoplastica was found in the trachea (case 2, Fig. -

Prenatal Diagnosis of a Huge Foetal Immature Sacrococcygeal Teratoma
Page 1 of 6 Case report Prenatal diagnosis of a huge foetal immature sacrococcygeal teratoma: our experience of a rare case and review Obstetrics & Gynecology of the literature M Varras1*, G Diakakis1, I Monselas1, CH Akrivis2 Abstract 6 - Introduction - increased blood flow into the tumour.- behaviour . Sacrococcygeal terato Pulsed Doppler showed the resist mas arise from the Hensen’s nodule Sacrococcygeal teratomas are rare- ance index of flow velocity wave- located on the anterior surface of the germ cell tumours associated with forms on the tumoural arteries to be- sacrum or coccyx11,12 and may be cystic, high perinatal and postnatal mor 0.51. Amnioreduction of 740 cc amni multilocular or cystic, and solid and tality and morbidity. The purpose of otic fluid was performed under ultra variable in size . our study was to report a case of a sonographic examination. Caesarean- The morbidity and mortality of foetal huge immature sacrococcygeal section was performed at 33 weeks’ the sacrococcygeal teratomas are low- teratoma diagnosed prenatally and- gestation due to profuse polyhydram when they are diagnosed prenatally8,9 managed successfully in the early nios via an upper vertical incision in and the surgical management is per Caseneonatal report period with surgical resec the uterus. After the stabilization of formed during the neonatal period . tion of the tumour. the newborn, tumour resection was- The prognosis is usually poor when successfully performed on the first they are diagnosed prior to 30 weeks’ A foetal mass with solid and× cystic day after delivery. Grossly, in the sur gestation and are predominantly solid components in the sacral region of- gical specimen the tumour measured with high vascularisation due to high- the foetus measuring 42 34 mm Conclusion18 cm in its maximum diameter and output cardiac failure, development- was detected during the foetal anat weighted 1 500 g.