Durham Research Online
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A 475 Years-Old Founder Effect Involving IL12RB1: a Highly Prevalent Mutation Conferring Mendelian Susceptibility to Mycobacterial Diseases in European Descendants
Infection, Genetics and Evolution 9 (2009) 574–580 Contents lists available at ScienceDirect Infection, Genetics and Evolution journal homepage: www.elsevier.com/locate/meegid A 475 years-old founder effect involving IL12RB1: A highly prevalent mutation conferring Mendelian Susceptibility to Mycobacterial Diseases in European descendants J. Yancoski a, C. Rocco a, A. Bernasconi a, M. Oleastro a, L. Bezrodnik b, C. Vra´tnica c, F. Haerynck d, S.D. Rosenzweig a,* a Hospital Nacional de Pediatrı´a J. P. Garrahan, Buenos Aires, Argentina b Hospital de Nin˜os Ricardo Gutierrez, Buenos Aires, Argentina c Hospital de Nin˜os Juan Pablo II, Corrientes, Argentina d Ghent University Hospital, Ghent, Belgium ARTICLE INFO ABSTRACT Article history: Mutations in IFNGR1, IFNGR2, IL12RB1, IL12B, STAT1 and NEMO result in a common clinical phenotype Received 11 December 2008 known as Mendelian Susceptibility to Mycobacterial Diseases (MSMD). Interleukin-12 receptor b1 (IL- Received in revised form 13 February 2009 12Rb1) deficiency is the most common genetic etiology for MSMD. Known mutations affecting IL12RB1 Accepted 16 February 2009 are recessively inherited and are associated with null response to both IL-12 and IL-23. Mutation IL12RB1 Available online 9 March 2009 1623_1624delinsTT was originally described in 5 families from European origin (2 from Germany; 1 from Cyprus, France and Belgium). Interestingly, this same mutation was found in an unexpectedly high Keywords: prevalence among IL-12Rb1 deficient patients in Argentina: 5-out-of-6 individuals born to unrelated Hot spot families carried this particular change. To determine whether mutation 1623_1624delinsTT represents a Salmonella Bacillus Calmette Guerin DNA mutational hotspot or a founder effect, 34 polymorphic markers internal or proximal to IL12RB1 Interferon gamma were studied in the Argentinean and the Belgian patients. -

NASP Polyclonal Antibody
For Research Use Only NASP Polyclonal antibody Catalog Number:11323-1-AP Featured Product 7 Publications www.ptgcn.com Catalog Number: GenBank Accession Number: Recommended Dilutions: Basic Information 11323-1-AP BC010105 WB 1:500-1:2400 Size: GeneID (NCBI): IP 0.5-4.0 ug for IP and 1:500-1:2000 700 μg/ml 4678 for WB IHC 1:20-1:200 Source: Full Name: IF 1:10-1:100 Rabbit nuclear autoantigenic sperm protein Isotype: (histone-binding) IgG Calculated MW: Purification Method: 788 aa, 85 kDa Antigen affinity purification Observed MW: Immunogen Catalog Number: 138 kDa AG1824 Applications Tested Applications: Positive Controls: IF, IHC, IP, WB, ELISA WB : mouse testis tissue; Cited Applications: IP : mouse testis tissue; IHC, WB IHC : human testis tissue; human lymphoma tissue Species Specificity: human, mouse, rat IF : MCF-7 cells; Cited Species: human, mouse, rat Note-IHC: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 NASP (nuclear autoantigenic sperm protein) is associated with DNA replication, cell proliferation and cell cycle Background Information progression through functioning as a Histone H1 binding protein that mediates histone transport to the nucleus. NASP had two isoforms, tNASP (testis type) and sNASP (somatic type). The tNASP was mainly expressed in testis, a variety of malignant tumors, stem cells and embryonic tissues, while sNASP existed in all somatic mitosis cells. This antibody can recognize both isoforms. Notable Publications Author Pubmed ID Journal Application Qing Yuan 25378924 Int J Nanomedicine WB Nishibu Takahiro T 23229118 Biosci Trends WB Sofie De Munter 28032891 FEBS Lett WB Storage: Storage Store at -20ºC. -

SFRS14 Antibody (Monoclonal) (M01) Mouse Monoclonal Antibody Raised Against a Partial Recombinant SFRS14
苏州工业园区双圩路9号1幢 邮 编 : 215000 电 话 : 0512-88856768 SFRS14 Antibody (monoclonal) (M01) Mouse monoclonal antibody raised against a partial recombinant SFRS14. Catalog # AT3845a Specification SFRS14 Antibody (monoclonal) (M01) - Product info Application WB, IHC, IF, E Primary Accession Q8IX01 Other Accession NM_014884 Reactivity Human Host mouse Clonality monoclonal Isotype IgG2a Kappa Clone Names 3C5 Calculated MW 120207 SFRS14 Antibody (monoclonal) (M01) - Additional info Gene ID 10147 Antibody Reactive Against Recombinant Protein.Western Blot detection against Other Names Immunogen (36.3 KDa) . SURP and G-patch domain-containing protein 2, Arginine/serine-rich-splicing factor 14, Splicing factor, arginine/serine-rich 14, SUGP2, KIAA0365, SFRS14 Target/Specificity SFRS14 (NP_055699, 579 a.a. ~ 674 a.a) partial recombinant protein with GST tag. MW of the GST tag alone is 26 KDa. Dilution WB~~1:500~1000 Format Clear, colorless solution in phosphate buffered saline, pH 7.2 . Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. SFRS14 monoclonal antibody (M01), Precautions clone 3C5 Western Blot analysis of SFRS14 Antibody (monoclonal) (M01) is for research use only SFRS14 expression in Hela S3 NE ( (Cat # and not for use in diagnostic or therapeutic procedures. AT3845a ) SFRS14 Antibody (monoclonal) (M01) - Protocols Provided below are standard protocols that you may find useful for product applications. • Western Blot • Blocking Peptides • Dot Blot • Immunohistochemistry • Immunofluorescence • Immunoprecipitation Immunoperoxidase of monoclonal • Flow Cytomety antibody to SFRS14 on formalin-fixed • Cell Culture paraffin-embedded human testis. SFRS14 Antibody (monoclonal) (M01) - Background [antibody concentration 3 ug/ml] This gene encodes a member of the arginine/serine-rich family of splicing factors. -

Molecular Evolution of NASP and Conserved Histone H3/H4 Transport Pathway Syed Nabeel-Shah1, Kanwal Ashraf2, Ronald E Pearlman2 and Jeffrey Fillingham1*
Nabeel-Shah et al. BMC Evolutionary Biology 2014, 14:139 http://www.biomedcentral.com/1471-2148/14/139 RESEARCH ARTICLE Open Access Molecular evolution of NASP and conserved histone H3/H4 transport pathway Syed Nabeel-Shah1, Kanwal Ashraf2, Ronald E Pearlman2 and Jeffrey Fillingham1* Abstract Background: NASP is an essential protein in mammals that functions in histone transport pathways and maintenance of a soluble reservoir of histones H3/H4. NASP has been studied exclusively in Opisthokonta lineages where some functional diversity has been reported. In humans, growing evidence implicates NASP miss-regulation in the development of a variety of cancers. Although a comprehensive phylogenetic analysis is lacking, NASP-family proteins that possess four TPR motifs are thought to be widely distributed across eukaryotes. Results: We characterize the molecular evolution of NASP by systematically identifying putative NASP orthologs across diverse eukaryotic lineages ranging from excavata to those of the crown group. We detect extensive silent divergence at the nucleotide level suggesting the presence of strong purifying selection acting at the protein level. We also observe a selection bias for high frequencies of acidic residues which we hypothesize is a consequence of their critical function(s), further indicating the role of functional constraints operating on NASP evolution. Our data indicate that TPR1 and TPR4 constitute the most rapidly evolving functional units of NASP and may account for the functional diversity observed among well characterized family members. We also show that NASP paralogs in ray-finned fish have different genomic environments with clear differences in their GC content and have undergone significant changes at the protein level suggesting functional diversification. -

1 Mutational Heterogeneity in Cancer Akash Kumar a Dissertation
Mutational Heterogeneity in Cancer Akash Kumar A dissertation Submitted in partial fulfillment of requirements for the degree of Doctor of Philosophy University of Washington 2014 June 5 Reading Committee: Jay Shendure Pete Nelson Mary Claire King Program Authorized to Offer Degree: Genome Sciences 1 University of Washington ABSTRACT Mutational Heterogeneity in Cancer Akash Kumar Chair of the Supervisory Committee: Associate Professor Jay Shendure Department of Genome Sciences Somatic mutation plays a key role in the formation and progression of cancer. Differences in mutation patterns likely explain much of the heterogeneity seen in prognosis and treatment response among patients. Recent advances in massively parallel sequencing have greatly expanded our capability to investigate somatic mutation. Genomic profiling of tumor biopsies could guide the administration of targeted therapeutics on the basis of the tumor’s collection of mutations. Central to the success of this approach is the general applicability of targeted therapies to a patient’s entire tumor burden. This requires a better understanding of the genomic heterogeneity present both within individual tumors (intratumoral) and amongst tumors from the same patient (intrapatient). My dissertation is broadly organized around investigating mutational heterogeneity in cancer. Three projects are discussed in detail: analysis of (1) interpatient and (2) intrapatient heterogeneity in men with disseminated prostate cancer, and (3) investigation of regional intratumoral heterogeneity in -

Mouse Nasp Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Nasp Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Nasp conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Nasp gene (NCBI Reference Sequence: NM_016777 ; Ensembl: ENSMUSG00000028693 ) is located on Mouse chromosome 4. 16 exons are identified, with the ATG start codon in exon 2 and the TAA stop codon in exon 16 (Transcript: ENSMUST00000030456). Exon 6~7 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Nasp gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP24-72F14 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a null mutation display embryonic lethality before implantation. Exon 6 starts from about 12.94% of the coding region. The knockout of Exon 6~7 will result in frameshift of the gene. The size of intron 5 for 5'-loxP site insertion: 2248 bp, and the size of intron 7 for 3'-loxP site insertion: 4639 bp. The size of effective cKO region: ~2179 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 5 6 7 16 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Nasp Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Genome Wide Association Study of Hippocampal Subfield Volume in PTSD Cases and Trauma-Exposed Controls
bioRxiv preprint doi: https://doi.org/10.1101/456988; this version posted October 30, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Hippocampal subfield GWAS in PTSD Revision date: October 18, 2018 Genome wide association study of hippocampal subfield volume in PTSD cases and trauma-exposed controls Rajendra A. Morey, M.D., M.S. a, b, c, Melanie E. Garrett, M.S. a, d, Jennifer S. Stevens, Ph.D. e, Emily Clarke, M.A. a, c, Courtney C. Haswell, M.S. a, c, Sanne J.H. van Rooij, Ph.D. e, Negar Fani, Ph.D. e, Adriana Lori, Ph.D. e, VA Mid-Atlantic MIRECC Workgroup†, Christine E. Marx, M.D. a, b, Jean C. Beckham, Ph.D. a, Gregory McCarthy, Ph.D. f, Michael A. Hauser, Ph.D. a, d, Allison E. Ashley-Koch, Ph.D. a, d †The VA Mid-Atlantic MIRECC Workgroup includes the following contributors: Mira Brancu, Ph.D., Patrick S. Calhoun, Ph.D., Eric Dedert, Ph.D., Eric B. Elbogen, Ph.D., John A. Fairbank, Ph.D., Robin A. Hurley, M.D., Jason D. Kilts, Ph.D., Nathan Kimbrel, Ph.D., Angela Kirby, M.S., Scott D. McDonald, Ph.D., Ph.D., Scott D. Moore, M.D., Ph.D., Jennifer C. Naylor, Ph.D., Jared Rowland, Ph.D., Cindy Swinkels, Ph.D., Steven T. Szabo, M.D., Ph.D., Katherine H. Taber, Ph.D., Larry A. Tupler, Ph.D., Elizabeth E. Van Voorhees, Ph.D., H. -

Open Data for Differential Network Analysis in Glioma
International Journal of Molecular Sciences Article Open Data for Differential Network Analysis in Glioma , Claire Jean-Quartier * y , Fleur Jeanquartier y and Andreas Holzinger Holzinger Group HCI-KDD, Institute for Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2/V, 8036 Graz, Austria; [email protected] (F.J.); [email protected] (A.H.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 27 October 2019; Accepted: 3 January 2020; Published: 15 January 2020 Abstract: The complexity of cancer diseases demands bioinformatic techniques and translational research based on big data and personalized medicine. Open data enables researchers to accelerate cancer studies, save resources and foster collaboration. Several tools and programming approaches are available for analyzing data, including annotation, clustering, comparison and extrapolation, merging, enrichment, functional association and statistics. We exploit openly available data via cancer gene expression analysis, we apply refinement as well as enrichment analysis via gene ontology and conclude with graph-based visualization of involved protein interaction networks as a basis for signaling. The different databases allowed for the construction of huge networks or specified ones consisting of high-confidence interactions only. Several genes associated to glioma were isolated via a network analysis from top hub nodes as well as from an outlier analysis. The latter approach highlights a mitogen-activated protein kinase next to a member of histondeacetylases and a protein phosphatase as genes uncommonly associated with glioma. Cluster analysis from top hub nodes lists several identified glioma-associated gene products to function within protein complexes, including epidermal growth factors as well as cell cycle proteins or RAS proto-oncogenes. -

Intrinsic Disorder in Tetratricopeptide Repeat Proteins
International Journal of Molecular Sciences Article Intrinsic Disorder in Tetratricopeptide Repeat Proteins 1, 1, 1, 2 Nathan W. Van Bibber y, Cornelia Haerle y, Roy Khalife y, Bin Xue and Vladimir N. Uversky 1,3,4,* 1 Department of Molecular Medicine Morsani College of Medicine, University of South Florida, 12901 Bruce B. Downs Blvd., Tampa, FL 33612, USA; [email protected] (N.W.V.B.); [email protected] (C.H.); [email protected] (R.K.) 2 Department of Cell Biology, Microbiology and Molecular Biology, School of Natural Sciences and Mathematics, College of Arts and Sciences, University of South Florida, Tampa, FL 33620, USA; [email protected] 3 USF Health Byrd Alzheimer’s Research Institute, Morsani College of Medicine, University of South Florida, 12901 Bruce B. Downs Blvd., Tampa, FL 33612, USA 4 Institute for Biological Instrumentation, Russian Academy of Sciences, Federal Research Center “Pushchino Scientific Center for Biological Research of the Russian Academy of Sciences”, 4 Institutskaya St., Pushchino, 142290 Moscow Region, Russia * Correspondence: [email protected]; Tel.: +1-813-974-5816; Fax: +1-813-974-7357 These authors contributed equally to this work. y Received: 21 April 2020; Accepted: 22 May 2020; Published: 25 May 2020 Abstract: Among the realm of repeat containing proteins that commonly serve as “scaffolds” promoting protein-protein interactions, there is a family of proteins containing between 2 and 20 tetratricopeptide repeats (TPRs), which are functional motifs consisting of 34 amino acids. The most distinguishing feature of TPR domains is their ability to stack continuously one upon the other, with these stacked repeats being able to affect interaction with binding partners either sequentially or in combination. -
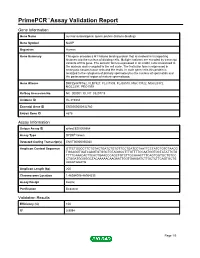
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name nuclear autoantigenic sperm protein (histone-binding) Gene Symbol NASP Organism Human Gene Summary This gene encodes a H1 histone binding protein that is involved in transporting histones into the nucleus of dividing cells. Multiple isoforms are encoded by transcript variants of this gene. The somatic form is expressed in all mitotic cells is localized to the nucleus and is coupled to the cell cycle. The testicular form is expressed in embryonic tissues tumor cells and the testis. In male germ cells this protein is localized to the cytoplasm of primary spermatocytes the nucleus of spermatids and the periacrosomal region of mature spermatozoa. Gene Aliases DKFZp547F162, FLB7527, FLJ31599, FLJ35510, MGC19722, MGC20372, MGC2297, PRO1999 RefSeq Accession No. NC_000001.10, NT_032977.9 UniGene ID Hs.319334 Ensembl Gene ID ENSG00000132780 Entrez Gene ID 4678 Assay Information Unique Assay ID qHsaCED0005564 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000350030 Amplicon Context Sequence GTTCTGGCCTTCTGTACTGATCTGTGTTCCTGATCCTAATTCCTATCTGTCTAACG TGGAGGTGATCAAGTGTGGCTGTAGGCCTTTGTTTTCCAATGGTGCTATATTCTG TTTTCAAACACTTCACTGAACCCAGCTGTCTTGCAAACTTTCAGTGGTGCTGTCC CTGGATGGGGGCTACAAAAACAAGAATTGGTGAAGATCTTGCTCTTCAGTGCTG AAAATGGATG Amplicon Length (bp) 200 Chromosome Location 1:46084006-46084235 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9994 Page 1/5 PrimePCR™Assay Validation Report cDNA Cq 20.64 cDNA Tm (Celsius) 83 gDNA Cq 22.63 Specificity -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -

Novel Targets of Apparently Idiopathic Male Infertility
International Journal of Molecular Sciences Review Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility Rossella Cannarella * , Rosita A. Condorelli , Laura M. Mongioì, Sandro La Vignera * and Aldo E. Calogero Department of Clinical and Experimental Medicine, University of Catania, 95123 Catania, Italy; [email protected] (R.A.C.); [email protected] (L.M.M.); [email protected] (A.E.C.) * Correspondence: [email protected] (R.C.); [email protected] (S.L.V.) Received: 8 February 2020; Accepted: 2 March 2020; Published: 3 March 2020 Abstract: Male infertility affects half of infertile couples and, currently, a relevant percentage of cases of male infertility is considered as idiopathic. Although the male contribution to human fertilization has traditionally been restricted to sperm DNA, current evidence suggest that a relevant number of sperm transcripts and proteins are involved in acrosome reactions, sperm-oocyte fusion and, once released into the oocyte, embryo growth and development. The aim of this review is to provide updated and comprehensive insight into the molecular biology of spermatogenesis, including evidence on spermatogenetic failure and underlining the role of the sperm-carried molecular factors involved in oocyte fertilization and embryo growth. This represents the first step in the identification of new possible diagnostic and, possibly, therapeutic markers in the field of apparently idiopathic male infertility. Keywords: spermatogenetic failure; embryo growth; male infertility; spermatogenesis; recurrent pregnancy loss; sperm proteome; DNA fragmentation; sperm transcriptome 1. Introduction Infertility is a widespread condition in industrialized countries, affecting up to 15% of couples of childbearing age [1]. It is defined as the inability to achieve conception after 1–2 years of unprotected sexual intercourse [2].