Transcription Factors Mix1 and Vegt, Relocalization of Vegt Mrna, and Conserved Endoderm and Dorsal Specification in Frogs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Thermal Adaptation of Amphibians in Tropical Mountains
Thermal adaptation of amphibians in tropical mountains. Consequences of global warming Adaptaciones térmicas de anfibios en montañas tropicales: consecuencias del calentamiento global Adaptacions tèrmiques d'amfibis en muntanyes tropicals: conseqüències de l'escalfament global Pol Pintanel Costa ADVERTIMENT. La consulta d’aquesta tesi queda condicionada a l’acceptació de les següents condicions d'ús: La difusió d’aquesta tesi per mitjà del servei TDX (www.tdx.cat) i a través del Dipòsit Digital de la UB (diposit.ub.edu) ha estat autoritzada pels titulars dels drets de propietat intel·lectual únicament per a usos privats emmarcats en activitats d’investigació i docència. No s’autoritza la seva reproducció amb finalitats de lucre ni la seva difusió i posada a disposició des d’un lloc aliè al servei TDX ni al Dipòsit Digital de la UB. No s’autoritza la presentació del seu contingut en una finestra o marc aliè a TDX o al Dipòsit Digital de la UB (framing). Aquesta reserva de drets afecta tant al resum de presentació de la tesi com als seus continguts. En la utilització o cita de parts de la tesi és obligat indicar el nom de la persona autora. ADVERTENCIA. La consulta de esta tesis queda condicionada a la aceptación de las siguientes condiciones de uso: La difusión de esta tesis por medio del servicio TDR (www.tdx.cat) y a través del Repositorio Digital de la UB (diposit.ub.edu) ha sido autorizada por los titulares de los derechos de propiedad intelectual únicamente para usos privados enmarcados en actividades de investigación y docencia. -
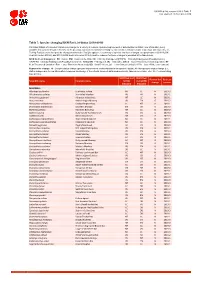
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

TESIS DGBARRAGAN.Pdf
PONTIFICIA UNIVERSIDAD CATÓLICA DEL ECUADOR FACULTAD DE CIENCIAS EXACTAS Y NATURALES ESCUELA DE CIENCIAS BIOLÓGICAS Effects of future climate change and habitat loss in the distribution of frog species in the Ecuadorian Andes Tesis previa a la obtención del título de Magíster en Biología de la Conservación DAYANA GABRIELA BARRAGÁN ALTAMIRANO Quito, 2015 iii Certifico que la tesis de Maestría en Biología de la Conservación de la candidata Dayana Gabriela Barragán Altamirano ha sido concluida de conformidad con las normas establecidas; por lo tanto, puede ser presentada para la calificación correspondiente. Firma de Director de Tesis Fecha: 27 diciembre de 2015 iv ACKNOWLEDGEMENTS This study has benefited from two valuable reviewers Veronica Crespo and Santiago Mena, who generously contributed with observations to improve this study. We also want to thank Santiago Burneo for guidance on the ecological niche modeling section, and Julio Sanchez, for his guidance on the section of statistical data analysis. We want to thank Damian Nicolalde for facilitating the data base of species occurrence records and Pablo Venegas for his assistance in taxonomic identification. We acknowledge Francisco Prieto, Danilo Granja, Janeth Delgado, Carlos Ponce (Ministerio del Ambiente), Alejandro Oviedo (Ministerio de Transporte y Obras Públicas), Pamela Hidalgo, and Francisco Moscoso who generously authorized and shared geographical information useful to perform the predictive habitat loss model. We want to thank Luis Camacho who helped to review the manuscript. Finally, Sean McCartney, Michelle Andrews (Clark Labs Technical Support), Santiago Silva (Centro de Recursos IDRISI Ecuador) offered their assistance during the predicting habitat loss modeling. v TABLE OF CONTENTS 1. -

Twenty-Fifth Meeting of the Animals Committee
AC25 Doc. 22 (Rev. 1) Annex 3 (English only / únicamente en inglés / seulement en anglais) Annex 3 Fauna: new species and other changes relating to species listed in the EC wildlife trade regulations – Report compiled by UNEP-WCMC to the European Commission, March, 2011 AC25 Doc. 22 (Rev. 1) Annex 3 – p. 1 Fauna: new species and other taxonomic changes relating to species listed in the EC wildlife trade regulations March, 2011 A report to the European Commission Directorate General E - Environment ENV.E.2. – Environmental Agreements and Trade by the United Nations Environment Programme World Conservation Monitoring Centre AC25 Doc. 22 (Rev. 1) Annex 3 – p. 2 UNEP World Conservation Monitoring Centre 219 Huntingdon Road Cambridge CB3 0DL United Kingdom Tel: +44 (0) 1223 277314 Fax: +44 (0) 1223 277136 Email: [email protected] Website: www.unep-wcmc.org CITATION UNEP-WCMC. 2011. Fauna: new species and other taxonomic changes relating to species ABOUT UNEP-WORLD CONSERVATION listed in the EC wildlife trade regulations. A MONITORING CENTRE report to the European Commission. UNEP- The UNEP World Conservation Monitoring WCMC, Cambridge. Centre (UNEP-WCMC), based in Cambridge, UK, is the specialist biodiversity information and assessment centre of the United Nations Environment Programme (UNEP), run PREPARED FOR cooperatively with WCMC, a UK charity. The The European Commission, Brussels, Belgium Centre's mission is to evaluate and highlight the many values of biodiversity and put authoritative biodiversity knowledge at the DISCLAIMER centre of decision-making. Through the analysis and synthesis of global biodiversity knowledge The contents of this report do not necessarily the Centre provides authoritative, strategic and reflect the views or policies of UNEP or timely information for conventions, countries contributory organisations. -

Notice to the Wildlife Import/Export Community
NOTICE TO THE WILDLIFE IMPORT/EXPORT COMMUNITY May 14, 2013 Subject: Changes to CITES Species Listings Background: Party countries of the Convention on International Trade in Endangered Species (CITES) meet approximately every two years for a Conference of the Parties. During these meetings, countries review and vote on amendments to the listings of protected species in CITES Appendix I and Appendix II. Such amendments become effective 90 days after the last day of the meeting unless Party countries agree to delay implementation. The most recent Conference of the Parties (CoP 16) was held in Bangkok, Thailand March 3-14, 2013. Action: The amendments to CITES Appendices I and II that appear below (which were adopted at CoP 16) will be effective on June 12, 2013, except for six listings of sharks and rays that have a delayed effective date of September 14, 2014. Any specimens of these species imported into, or exported from, the United States on or after June 12, 2013 (or September 14, 2014 for the six shark/ray listings) will require CITES documentation as specified under the amended listings. The import, export, or re-export of shipments of these species that are accompanied by CITES documents reflecting a pre-June 12 (or September 14 2014 for the six shark/ray listings) listing status or that lack CITES documents because no listing was previously in effect must be completed by midnight (local time at the point of import/export) on June 11, 2013 (or September 13, 2014 for the six shark/ray listings). Importers and exporters can find the official revised CITES appendices on the CITES website at http://www.cites.org. -

Jama-Coaque Reserve VCA Proposal
1. EXECUTIVE SUMMARY Organization: Third Millennium Alliance (USA)/ Grupo Ecológico Jama-Coaque (Ecuador) Project Name: Reforestation, Conservation, and Community Outreach in Coastal Ecuador Proposed VCA: The Jama-Coaque Reserve Location: Coastal Ecuador’s Pacific Forests: Province of Manabi, Ecuador. Mission: To preserve the last remnants of coastal Ecuador's Pacific Forest and to empower local communities to restore what has been lost Vision: We envision a culture in which local communities recognize both the practical and intrinsic benefits of forest stewardship and manage the land accordingly, creating both economic and environmental sustainability in the region Contact Person: Ryan L. Lynch, Executive Director; Jose Tamayo 1024 y Lizardo Garcia, Quito, Ecuador; Phone: +593 98 732 5336; Email: [email protected]; Website: www.tmalliance.org Project Overview: Named after the Pre-Incan Jama Coaque culture that inhabited the area, the Jama-Coaque Reserve currently protects 411 hectares of tropical moist forest and premontane cloud forest in one of the last major remnants of Pacific Equatorial Forest in the Tumbes-Chocó transition zone along the coastal mountain range of northwest Ecuador. The region surrounding the Reserve has seen one of the highest rates of deforestation in South America, with only 2% of native forest cover remaining. The strategic location of the Reserve along the coastal Manabi mountain range protects the headwaters of two major river systems, which provide water to the community of Camarones and protect the habitats of a variety of rare, endemic, and globally threatened species. To date, 255 species of birds (16 globally threatened), 41 species of reptile (18 globally threatened), 27 species of amphibian (5 globally threatened, plus 3+ species new to science), and 18 species of mammals (2 globally threatened) have been documented in the Reserve. -

Diversity Within Diversity Parasite Species Richness in Poison Frogs
Molecular Phylogenetics and Evolution 125 (2018) 40–50 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Diversity within diversity: Parasite species richness in poison frogs assessed T by transcriptomics ⁎ Juan C. Santosa, , Rebecca D. Tarvinb, Lauren A. O'Connellc, David C. Blackburnd, Luis A. Colomae,f a Department of Biological Sciences, St. John's University, United States b Integrative Biology, University of Texas at Austin, United States c Department of Biology, Stanford University, United States d Florida Museum of Natural History, United States e Centro Jambatu de Investigación y Conservación de Anfibios, Fundación Otonga, Ecuador f Universidad Regional Amazónica Ikiam, Ecuador ARTICLE INFO ABSTRACT Keywords: Symbionts (e.g., endoparasites and commensals) play an integral role in their host's ecology, yet in many cases Transcriptomics their diversity is likely underestimated. Although endoparasites are traditionally characterized using mor- Co-diversification phology, sequences of conserved genes, and shotgun metagenomics, host transcriptomes constitute an underused Amphibians resource to identify these organisms’ diversity. By isolating non-host transcripts from host transcriptomes, in- Aposematism dividual host tissues can now simultaneously reveal their endoparasite species richness (i.e., number of different Parasitology taxa) and provide insights into parasite gene expression. These approaches can be used in host taxa whose endoparasites are mostly unknown, such as those of tropical amphibians. Here, we focus on the poison frogs (Dendrobatidae) as hosts, which are a Neotropical clade known for their bright coloration and defensive alka- loids. These toxins are an effective protection against vertebrate predators (e.g., snakes and birds), bacteria, and skin-biting ectoparasites (e.g., mosquitoes); however, little is known about their deterrence against eukaryotic endoparasites. -

IACUC Update
IACUC Update IACUCInstitutional Animal Care and Use Committee Update The University of Texas at Austin July 2014 Volume 7, Issue 10 “Introduction to Wildlife” offered Meeting Requirements for in the AALAS Learning Library Alternative Searches: Webinar Do you conduct studies with wildlife in Available for Viewing research? If so, there is a new AALAS A recording of the OLAW Online Seminar Learning Library (ALL) module “Introduction “Meeting Requirements for Alternatives to Wildlife.” This module will provide Searches: Information for IACUCs and information on: Investigators” broadcast on June 26, 2014, Protocol review issues has been posted on the OLAW website.- Lorem Ipsum • • Occupational health issues In this webinar, Kathleen Gregory, reference • Transportation and holding of wild librarian at the University of Denver, caught animals provided information for IACUCs and • Capture methods researchers about the alternatives search • Marking or identification methods required by the USDA Animal Welfare • Proper permits from local and federal Regulations. agencies The recording and supporting materials can • Recognition and management of pain be found on the Education Resources and distress in wild animals webpage. • Euthanasia, and more! When you view the webinar, please email At the July 14, 2014 Full Committee Review, Justin McNulty ([email protected]) the IACUC voted to require this module for all so that your TXClass training record can be non-fish, non-amphibian, non-traditional documented. laboratory animal users, effective December 1, 2014. For a specific listing of affected species, please refer to the attached listing. eProtocol Tip of the Month On December 1, 2014, all species listed in the Remember: Protocols, Amendments, and attached listing will be coded in eProtocol for Continuing Review applications cannot be this course. -
ANÁLISIS PARA EL DICTAMEN DE EXTRACCIÓN NO PERJUDICIAL DE Epipedobates Anthonyi Y Epipedobates Tricolor
ANÁLISIS PARA EL DICTAMEN DE EXTRACCIÓN NO PERJUDICIAL DE Epipedobates anthonyi Y Epipedobates tricolor Solicitud presentada por: WIKIRI S.A. Preparado para la Facultad de Ciencias Naturales de la Universidad de Guayaquil Autoridad Científica CITES Informe elaborado por: Luis A. Amador O. GUAYAQUIL, FEBRERO 2014 Tabla de contenido 1 INTRODUCCIÓN .............................................................................................................. 2 1.1 Antecedentes ............................................................................................................... 2 1.2 Anfibios de Ecuador .................................................................................................. 2 1.3 Comercio de anfibios ................................................................................................. 3 2 DICTAMEN DE EXTRACCIÓN NO PERJUDICIAL DE LA CITES........................... 4 3 EVALUACIÓN DE DICTAMEN DE EXTRACCIÓN NO PERJUDICIAL ................ 6 3.1 Metodología ................................................................................................................ 6 3.2 Especies ........................................................................................................................ 8 3.2.1 Epipedobates anthonyi, Noble (1921) .................................................................. 9 3.2.2 Epipedobates tricolor, Boulenger (1899) ........................................................... 13 4 CUPOS .............................................................................................................................. -

Variation in the Schedules of Somite and Neural Development in Frogs
Variation in the schedules of somite and neural development in frogs Natalia Sáenz-Poncea, Christian Mitgutschb, and Eugenia M. del Pinoa,1 aEscuela de Ciencias Biológicas, Pontificia Universidad Católica del Ecuador, Quito 17, Ecuador; and bLaboratory for Evolutionary Morphology, RIKEN Center for Developmental Biology, Kobe 650-0047 Hyogo, Japan Contributed by Eugenia M. del Pino, November 7, 2012 (sent for review September 3, 2012) The timing of notochord, somite, and neural development was riobambae, the developing embryos are transported in a pouch of analyzed in the embryos of six different frog species, which have integument by the mother (14, 15). The analyzed frogs belong to been divided into two groups, according to their developmental four different families and have notable differences in egg size speed. Rapid developing species investigated were Xenopus laevis (Table 1). Frog species that require hours to advance from fertil- (Pipidae), Engystomops coloradorum,andEngystomops randi (Leiu- ization to the end of gastrulation are considered to develop rap- peridae). The slow developers were Epipedobates machalilla and idly, whereas those that take days for the same process are Epipedobates tricolor (Dendrobatidae) and Gastrotheca riobambae considered to develop slowly (Table 1). Differences in rates of (Hemiphractidae). Blastopore closure, notochord formation, somite development were also detected in advanced embryos of these development, neural tube closure, and the formation of cranial neu- frogs (13, 15, 16). In the rapidly developing embryos of E. colo- ral crest cell-streams were detected by light and scanning electron radorum and E. randi, elongation of the notochord was detected at microscopy and by immuno-histochemical detection of somite and the mid-gastrula stage, as in the embryos of X. -

Extended Version of the Federal Register Notice
ANNOUNCEMENT OF TENTATIVE U.S. NEGOTIATING POSITIONS FOR AGENDA ITEMS AND SPECIES PROPOSALS SUBMITTED BY FOREIGN GOVERNMENTS AND THE CITES SECRETARIAT We, the United States, as a Party to the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), will attend the sixteenth regular meeting of the Conference of the Parties to CITES (CoP16) in Bangkok, Thailand, during March 3–14, 2013. This notice announces the tentative U.S. negotiating positions on amendments to the CITES Appendices (species proposals), draft resolutions and decisions, and agenda items submitted by other countries and the CITES Secretariat for consideration at CoP16. Please note that we published in the Federal Register on February 28, 2013 the availability on our website of our tentative U.S. negotiating positions on amendments to the CITES Appendices (species proposals), draft resolutions and decisions, and agenda items submitted by other countries and the CITES Secretariat for consideration at CoP16. DATES: In further developing U.S. negotiating positions on these issues, we will continue to consider information and comments submitted in response to our notice of November 9, 2012 (77 FR 67390). We will also continue to consider information received at the public meeting (announced with a revised date in the Federal Register; 77 FR 71012), which was held on December 13, 2012. 1 Background The Convention on International Trade in Endangered Species of Wild Fauna and Flora, hereinafter referred to as CITES or the Convention, is an international treaty designed to control and regulate international trade in certain animal and plant species that are now or potentially may become threatened with extinction. -

Development and Gastrulation in Hyloxalus Vertebralis and Dendrobates Auratus (Anura: Dendrobatidae)
Official journal website: Amphibian & Reptile Conservation amphibian-reptile-conservation.org 8(1) [Special Section]: 121–135 (e90). Development and gastrulation in Hyloxalus vertebralis and Dendrobates auratus (Anura: Dendrobatidae) Francisca Hervas, Karina P. Torres, Paola Montenegro-Larrea, and 1Eugenia M. del Pino Escuela de Ciencias Biológicas, Pontificia Universidad Católica del Ecuador, Av. 12 de Octubre 1076 y Roca, Quito 170517, ECUADOR Abstract.—We document the embryonic development of Hyloxalus vertebralis, a frog species of the Ecuadorian highlands, declared as Critically Endangered by the International Union for the Conservation of Nature (IUCN) due to significant declines in its populations. Our work may be of value for conservation and management of this endangered frog, especially as it is being bred in captivity to ensure against extinction. We were able to analyze and compare the development of H. vertebralis with Dendrobates auratus (Dendrobatidae), and other frogs, because of the successful reproduction in captivity of Ecuadorian frogs at the Balsa de los Sapos, Centre of Amphibian Investigation and Conservation (CICA), of the Pontificia Universidad Católica del Ecuador, in Quito. Embryos were fixed, and the external and internal morphology was described from whole mounts, and serial sections. Cellular morphology was analyzed by staining nuclei. Embryos of H. vertebralis and D. auratus developed from eggs that were 2.6 and 3.5 mm in diameter, respectively. In spite of the large size of their eggs, the morphology of H. vertebralis embryos from cleavage to hatching was similar to the morphology of Epipedobates machalilla (Dendrobatidae) embryos. The comparison of gastrulation morphology was extended to six additional species of Dendrobatidae (E.