HNRNPA1 Promotes Recognition of Splice Site Decoys by U2AF2 In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Whole Exome Sequencing in ADHD Trios from Single and Multi-Incident Families Implicates New Candidate Genes and Highlights Polygenic Transmission
European Journal of Human Genetics (2020) 28:1098–1110 https://doi.org/10.1038/s41431-020-0619-7 ARTICLE Whole exome sequencing in ADHD trios from single and multi-incident families implicates new candidate genes and highlights polygenic transmission 1,2 1 1,2 1,3 1 Bashayer R. Al-Mubarak ● Aisha Omar ● Batoul Baz ● Basma Al-Abdulaziz ● Amna I. Magrashi ● 1,2 2 2,4 2,4,5 6 Eman Al-Yemni ● Amjad Jabaan ● Dorota Monies ● Mohamed Abouelhoda ● Dejene Abebe ● 7 1,2 Mohammad Ghaziuddin ● Nada A. Al-Tassan Received: 26 September 2019 / Revised: 26 February 2020 / Accepted: 10 March 2020 / Published online: 1 April 2020 © The Author(s) 2020. This article is published with open access Abstract Several types of genetic alterations occurring at numerous loci have been described in attention deficit hyperactivity disorder (ADHD). However, the role of rare single nucleotide variants (SNVs) remains under investigated. Here, we sought to identify rare SNVs with predicted deleterious effect that may contribute to ADHD risk. We chose to study ADHD families (including multi-incident) from a population with a high rate of consanguinity in which genetic risk factors tend to 1234567890();,: 1234567890();,: accumulate and therefore increasing the chance of detecting risk alleles. We employed whole exome sequencing (WES) to interrogate the entire coding region of 16 trios with ADHD. We also performed enrichment analysis on our final list of genes to identify the overrepresented biological processes. A total of 32 rare variants with predicted damaging effect were identified in 31 genes. At least two variants were detected per proband, most of which were not exclusive to the affected individuals. -

Congenital Disorders of Glycosylation from a Neurological Perspective
brain sciences Review Congenital Disorders of Glycosylation from a Neurological Perspective Justyna Paprocka 1,* , Aleksandra Jezela-Stanek 2 , Anna Tylki-Szyma´nska 3 and Stephanie Grunewald 4 1 Department of Pediatric Neurology, Faculty of Medical Science in Katowice, Medical University of Silesia, 40-752 Katowice, Poland 2 Department of Genetics and Clinical Immunology, National Institute of Tuberculosis and Lung Diseases, 01-138 Warsaw, Poland; [email protected] 3 Department of Pediatrics, Nutrition and Metabolic Diseases, The Children’s Memorial Health Institute, W 04-730 Warsaw, Poland; [email protected] 4 NIHR Biomedical Research Center (BRC), Metabolic Unit, Great Ormond Street Hospital and Institute of Child Health, University College London, London SE1 9RT, UK; [email protected] * Correspondence: [email protected]; Tel.: +48-606-415-888 Abstract: Most plasma proteins, cell membrane proteins and other proteins are glycoproteins with sugar chains attached to the polypeptide-glycans. Glycosylation is the main element of the post- translational transformation of most human proteins. Since glycosylation processes are necessary for many different biological processes, patients present a diverse spectrum of phenotypes and severity of symptoms. The most frequently observed neurological symptoms in congenital disorders of glycosylation (CDG) are: epilepsy, intellectual disability, myopathies, neuropathies and stroke-like episodes. Epilepsy is seen in many CDG subtypes and particularly present in the case of mutations -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology. -
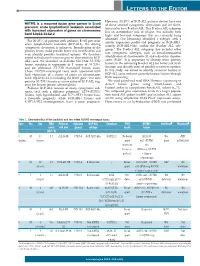
NUTM1 Is a Recurrent Fusion Gene Partner in B-Cell Precursor Acute
LETTERS TO THE EDITOR However, 20-25% of BCP-ALL patients do not have one NUTM1 is a recurrent fusion gene partner in B-cell of these sentinel cytogenetic aberrations and are there- precursor acute lymphoblastic leukemia associated fore said to have B-other ALL. This B-other ALL subgroup with increased expression of genes on chromosome has an intermediate risk of relapse, but includes both band 10p12.31-12.2 high- and low-risk subgroups that are currently being identified. Our laboratory identified a subtype with a For 20-25% of patients with pediatric B-cell precursor similar expression profile and prognosis as BCR-ABL1, acute lymphoblastic leukemia (BCP-ALL), the driving namely BCR-ABL1-like, within the B-other ALL sub- cytogenetic aberration is unknown. Identification of the group.2 The B-other ALL subgroup also includes other primary lesion could provide better risk stratification and rare cytogenetic subtypes, such as intrachromosomal even identify possible treatment options. We therefore amplification of chromosome 21 and a dicentric chromo- aimed to find novel recurrent genetic aberrations in BCP- 1 ALL cases. We identified an in-frame SLC12A6-NUTM1 some (9;20). It is important to identify more primary fusion, resulting in expression of 3’ exons of NUTM1, lesions in the remaining B-other ALL for better risk strat- and six additional NUTM1-rearranged fusion cases. ification and identification of possible treatment options. These NUTM1-rearranged cases were associated with In this study, we aimed to identify recurrent fusions in high expression of a cluster of genes on chromosome BCP-ALL cases without currently known lesions through band 10p12.31-12.2, including the BMI1 gene. -

Revostmm Vol 10-4-2018 Ingles Maquetaciûn 1
108 ORIGINALS / Rev Osteoporos Metab Miner. 2018;10(4):108-18 Roca-Ayats N1, Falcó-Mascaró M1, García-Giralt N2, Cozar M1, Abril JF3, Quesada-Gómez JM4, Prieto-Alhambra D5,6, Nogués X2, Mellibovsky L2, Díez-Pérez A2, Grinberg D1, Balcells S1 1 Departamento de Genética, Microbiología y Estadística - Facultad de Biología - Universidad de Barcelona - Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER) - Instituto de Salud Carlos III (ISCIII) - Instituto de Biomedicina de la Universidad de Barcelona (IBUB) - Instituto de Investigación Sant Joan de Déu (IRSJD) - Barcelona (España) 2 Unidad de Investigación en Fisiopatología Ósea y Articular (URFOA); Instituto Hospital del Mar de Investigaciones Médicas (IMIM) - Parque de Salud Mar - Centro de Investigación Biomédica en Red de Fragilidad y Envejecimiento Saludable (CIBERFES); Instituto de Salud Carlos III (ISCIII) - Barcelona (España) 3 Departamento de Genética, Microbiología y Estadística; Facultad de Biología; Universidad de Barcelona - Instituto de Biomedicina de la Universidad de Barcelona (IBUB) - Barcelona (España) 4 Unidad de Metabolismo Mineral; Instituto Maimónides de Investigación Biomédica de Córdoba (IMIBIC); Hospital Universitario Reina Sofía - Centro de Investigación Biomédica en Red de Fragilidad y Envejecimiento Saludable (CIBERFES); Instituto de Salud Carlos III (ISCIII) - Córdoba (España) 5 Grupo de Investigación en Enfermedades Prevalentes del Aparato Locomotor (GREMPAL) - Instituto de Investigación en Atención Primaria (IDIAP) Jordi Gol - Centro de Investigación -

Distinct Splicing Signatures Affect Converged Pathways in Myelodysplastic Syndrome Patients Carrying Mutations in Different Splicing Regulators
Downloaded from rnajournal.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators JINSONG QIU,1,7 BING ZHOU,1,7 FELICITAS THOL,2 YU ZHOU,1 LIANG CHEN,1 CHANGWEI SHAO,1 CHRISTOPHER DEBOEVER,3 JIAYI HOU,4 HAIRI LI,1 ANUHAR CHATURVEDI,2 ARNOLD GANSER,2 RAFAEL BEJAR,5 DONG-ER ZHANG,6 XIANG-DONG FU,1,3 and MICHAEL HEUSER2 1Department of Cellular and Molecular Medicine, School of Medicine, University of California, San Diego, La Jolla, California 92093, USA 2Department of Hematology, Hemostasis, Oncology and Stem cell Transplantation, Hannover Medical School, 30625 Hannover, Germany 3Institute for Genomic Medicine, University of California, San Diego, La Jolla, California 92093, USA 4Clinical and Translational Research Institute, University of California, San Diego, La Jolla, California 92093, USA 5Division of Hematology-Oncology, Moores Cancer Center, University of California, San Diego, La Jolla, California 92093, USA 6Department of Pathology, Moores Cancer Center, University of California, San Diego, La Jolla, California 92093, USA ABSTRACT Myelodysplastic syndromes (MDS) are heterogeneous myeloid disorders with prevalent mutations in several splicing factors, but the splicing programs linked to specific mutations or MDS in general remain to be systematically defined. We applied RASL-seq, a sensitive and cost-effective platform, to interrogate 5502 annotated splicing events in 169 samples from MDS patients or healthy individuals. We found that splicing signatures associated with normal hematopoietic lineages are largely related to cell signaling and differentiation programs, whereas MDS-linked signatures are primarily involved in cell cycle control and DNA damage responses. -

Viewed and Published Immediately Upon Acceptance Cited in Pubmed and Archived on Pubmed Central Yours — You Keep the Copyright
BMC Genomics BioMed Central Research article Open Access Differential gene expression in ADAM10 and mutant ADAM10 transgenic mice Claudia Prinzen1, Dietrich Trümbach2, Wolfgang Wurst2, Kristina Endres1, Rolf Postina1 and Falk Fahrenholz*1 Address: 1Johannes Gutenberg-University, Institute of Biochemistry, Mainz, Johann-Joachim-Becherweg 30, 55128 Mainz, Germany and 2Helmholtz Zentrum München – German Research Center for Environmental Health, Institute for Developmental Genetics, Ingolstädter Landstraße 1, 85764 Neuherberg, Germany Email: Claudia Prinzen - [email protected]; Dietrich Trümbach - [email protected]; Wolfgang Wurst - [email protected]; Kristina Endres - [email protected]; Rolf Postina - [email protected]; Falk Fahrenholz* - [email protected] * Corresponding author Published: 5 February 2009 Received: 19 June 2008 Accepted: 5 February 2009 BMC Genomics 2009, 10:66 doi:10.1186/1471-2164-10-66 This article is available from: http://www.biomedcentral.com/1471-2164/10/66 © 2009 Prinzen et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: In a transgenic mouse model of Alzheimer disease (AD), cleavage of the amyloid precursor protein (APP) by the α-secretase ADAM10 prevented amyloid plaque formation, and alleviated cognitive deficits. Furthermore, ADAM10 overexpression increased the cortical synaptogenesis. These results suggest that upregulation of ADAM10 in the brain has beneficial effects on AD pathology. Results: To assess the influence of ADAM10 on the gene expression profile in the brain, we performed a microarray analysis using RNA isolated from brains of five months old mice overexpressing either the α-secretase ADAM10, or a dominant-negative mutant (dn) of this enzyme. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Open Data for Differential Network Analysis in Glioma
International Journal of Molecular Sciences Article Open Data for Differential Network Analysis in Glioma , Claire Jean-Quartier * y , Fleur Jeanquartier y and Andreas Holzinger Holzinger Group HCI-KDD, Institute for Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2/V, 8036 Graz, Austria; [email protected] (F.J.); [email protected] (A.H.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 27 October 2019; Accepted: 3 January 2020; Published: 15 January 2020 Abstract: The complexity of cancer diseases demands bioinformatic techniques and translational research based on big data and personalized medicine. Open data enables researchers to accelerate cancer studies, save resources and foster collaboration. Several tools and programming approaches are available for analyzing data, including annotation, clustering, comparison and extrapolation, merging, enrichment, functional association and statistics. We exploit openly available data via cancer gene expression analysis, we apply refinement as well as enrichment analysis via gene ontology and conclude with graph-based visualization of involved protein interaction networks as a basis for signaling. The different databases allowed for the construction of huge networks or specified ones consisting of high-confidence interactions only. Several genes associated to glioma were isolated via a network analysis from top hub nodes as well as from an outlier analysis. The latter approach highlights a mitogen-activated protein kinase next to a member of histondeacetylases and a protein phosphatase as genes uncommonly associated with glioma. Cluster analysis from top hub nodes lists several identified glioma-associated gene products to function within protein complexes, including epidermal growth factors as well as cell cycle proteins or RAS proto-oncogenes. -

Genetic Studies of Human Hereditary Skin Disorders: Ectodermal Dysplasias and Alopecias
Genetic Studies of Human Hereditary Skin Disorders: Ectodermal Dysplasias and Alopecias by Muhammad Tariq Department of Biochemistry Faculty of Biological Sciences Quaid-I-Azam University Islamabad, Pakistan 2009 Genetic Studies of Human Hereditary Skin Disorders: Ectodermal Dysplasias and Alopecias A thesis submitted in the partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biochemistry/ Molecular Biology by Muhammad Tariq Department of Biochemistry Faculty of Biological Sciences Quaid-I-Azam University Islamabad, Pakistan 2009 In the name of Allah, the Most Gracious, the Most Merciful He is the One who created the heavens and the earth, truthfully. Whenever He says, "Be," it is. His word is the absolute truth. All sovereignty belongs to Him the day the horn is blown. Knower of all secrets and declarations, He is the Most Wise, the Cognizant. (Al-Quran 6:73) Dedicated to My sweet & beloved parents & brothers Declaration I hereby declared that the work presented in this thesis is my own effort and hard work and it is written and composed by me. No part of this thesis has been previously published or presented for any other degree or certificate. Muhammad Tariq Contents CONTENTS Title Page No. Preface Acknowledgements I List of Abbreviations III List of Tables VII List of Figures XI Summary XIX List of Publications XXIII Chapter 1: Introduction 1 Human Skin 1 Epidermis 2 Dermis 2 Hypodermis 2 Ectodermal Appendages 2 Hair 2 Nail 3 Tooth 4 Sweat Glands 5 Genetic Studies of Human Hereditary Skin -

Molecular Mechanisms in Muscular Dystrophy : a Gene Expression Profiling Study Turk, R
Molecular mechanisms in muscular dystrophy : a gene expression profiling study Turk, R. Citation Turk, R. (2006, September 27). Molecular mechanisms in muscular dystrophy : a gene expression profiling study. Retrieved from https://hdl.handle.net/1887/4577 Version: Corrected Publisher’s Version Licence agreement concerning inclusion of doctoral License: thesis in the Institutional Repository of the University of Leiden Downloaded from: https://hdl.handle.net/1887/4577 Note: To cite this publication please use the final published version (if applicable). Molecular Mechanisms In Muscular Dystrophy A Gene Expression Profiling Study Molecular Mechanisms In Muscular Dystrophy A Gene Expression Profiling Study Proefschrift ter verkrijging van de graad van Doctor aan de Universiteit Leiden, op gezag van de Rector Magnificus Dr. D.D.Breimer, hoogleraar in de faculteit der Wiskunde en Natuurwetenschappen en die der Geneeskunde, volgens besluit van het College voor Promoties te verdedigen op woensdag 27 september 2006 klokke 15.00 uur door Rolf Turk Geboren te Leiden in 1975 Promotiecommissie Promotor Prof. Dr. G.J.B. van Ommen Co-promotores Dr. J.T. den Dunnen Dr. P.A.C. ‘t Hoen Referent Prof. Dr. R.M.W. Hofstra (Rijksuniversiteit Groningen) Overige leden Prof. Dr. M. Koenig (Université Louis Pasteur de Strasbourg) An experiment is a question which science poses to Nature, and a measurement is the recording of Nature’s answer. Max Planck Aan Maaike, Gerard en Annet Printed by: Drukkerij Duineveld ISBN-10: 90-9021042-3 ISBN-13: 978-90-9021042-1 Turk, Rolf Molecular mechanisms in muscular dystrophy. A gene expression profiling study. Thesis, Leiden University Medical Center September 27, 2006 © Rolf Turk No part of this thesis may be reproduced or transmitted in any form or by any means, without the written permission of the copyright owner Molecular Mechanisms In Muscular Dystrophies Preface 9 Chapter 1 Introduction 11 1. -

ARP55242 P050) Data Sheet
COG4 antibody - middle region (ARP55242_P050) Data Sheet Product Number ARP55242_P050 Product Name COG4 antibody - middle region (ARP55242_P050) Size 50ug Gene Symbol COG4 Alias Symbols COD1; DKFZp586E1519; CDG2J Nucleotide Accession# NM_015386 Protein Size (# AA) 789 amino acids Molecular Weight 89kDa Subunit 4 Product Format Lyophilized powder NCBI Gene Id 25839 Host Rabbit Clonality Polyclonal Official Gene Full Name Component of oligomeric golgi complex 4 Gene Family COG This is a rabbit polyclonal antibody against COG4. It was validated on Western Blot using a cell lysate as a Description positive control. Aviva Systems Biology strives to provide antibodies covering each member of a whole protein family of your interest. We also use our best efforts to provide you antibodies recognize various epitopes of a target protein. For availability of antibody needed for your experiment, please inquire (). Peptide Sequence Synthetic peptide located within the following region: LFSQGIGGEQAQAKFDSCLSDLAAVSNKFRDLLQEGLTELNSTAIKPQVQ Target Reference Suzuki,Y., (2004) Genome Res. 14 (9), 1711-1718 Multiprotein complexes are key determinants of Golgi apparatus structure and its capacity for intracellular transport and glycoprotein modification. Several complexes have been identified, including the Golgi transport complex (GTC), the LDLC complex, which is involved in glycosylation reactions, and the SEC34 complex, which is involved in vesicular transport. These 3 complexes are identical and have been termed the conserved oligomeric Golgi (COG) complex, which includes COG4.Multiprotein complexes are key determinants of Golgi Description of Target apparatus structure and its capacity for intracellular transport and glycoprotein modification. Several complexes have been identified, including the Golgi transport complex (GTC), the LDLC complex, which is involved in glycosylation reactions, and the SEC34 complex, which is involved in vesicular transport.