Form and Structure Factors: Modeling and Interactions Jan Skov Pedersen, Department of Chemistry and Inano Center University of Aarhus Denmark SAXS Lab
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthesis, Rheology, Neutron Scattering
Downloaded from orbit.dtu.dk on: May 16, 2018 Entangled Polymer Melts in Extensional Flow: Synthesis, Rheology, Neutron Scattering Dorokhin, Andriy Publication date: 2018 Document Version Publisher's PDF, also known as Version of record Link back to DTU Orbit Citation (APA): Dorokhin, A. (2018). Entangled Polymer Melts in Extensional Flow: Synthesis, Rheology, Neutron Scattering. DTU Nanotech. General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Entangled Polymer Melts in Extensional Flow: Synthesis, Rheology, Neutron Scattering Andriy Dorokhin PhD Thesis December 2017 Ph.D. Thesis Doctor of Philosophy Entangled Polymer Melts in Extensional Flow: Synthesis, Rheology, Neutron Scattering Andriy Dorokhin Kongens Lyngby 2018 DTU Nanotech Department of Micro- and Nanotechnology Technical University of Denmark Produktionstorvet, -
Structure Factors Again
Structure factors again • Remember 1D, structure factor for order h 1 –Fh = |Fh|exp[iαh] = I0 ρ(x)exp[2πihx]dx – Where x is fractional position along “unit cell” distance (repeating distance, origin arbitrary) • As if scattering was coming from a single “center of gravity” scattering point • Presence of “h” in equation means that structure factors of different orders have different phases • Note that exp[2πihx]dx looks (and behaves) like a frequency, but it’s not (dx has to do with unit cell, and the sum gives the phase Back and Forth • Fourier sez – For any function f(x), there is a “transform” of it which is –F(h) = Ûf(x)exp(2pi(hx))dx – Where h is reciprocal of x (1/x) – Structure factors look like that • And it works backward –f(x) = ÛF(h)exp(-2pi(hx))dh – Or, if h comes only in discrete points –f(x) = SF(h)exp(-2pi(hx)) Structure factors, cont'd • Structure factors are a "Fourier transform" - a sum of components • Fourier transforms are reversible – From summing distribution of ρ(x), get hth order of diffraction – From summing hth orders of diffraction, get back ρ(x) = Σ Fh exp[-2πihx] Two dimensional scattering • In Frauenhofer diffraction (1D), we considered scattering from points, along the line • In 2D diffraction, scattering would occur from lines. • Numbering of the lines by where they cut the edges of a unit cell • Atom density in various lines can differ • Reflections now from planes Extension to 3D – Planes defined by extension from 2D case – Unit cells differ • Depends on arrangement of materials in 3D lattice • = "Space -

System Design and Verification of the Precession Electron Diffraction Technique
NORTHWESTERN UNIVERSITY System Design and Verification of the Precession Electron Diffraction Technique A DISSERTATION SUBMITTED TO THE GRADUATE SCHOOL IN PARTIAL FULFILLMENT OF THE REQUIREMENTS for the degree DOCTOR OF PHILOSOPHY Field of Materials Science and Engineering By Christopher Su-Yan Own EVANSTON, ILLINOIS First published on the WWW 01, August 2005 Build 05.12.07. PDF available for download at: http://www.numis.northwestern.edu/Research/Current/precession.shtml c Copyright by Christopher Su-Yan Own 2005 All Rights Reserved ii ABSTRACT System Design and Verification of the Precession Electron Diffraction Technique Christopher Su-Yan Own Bulk structural crystallography is generally a two-part process wherein a rough starting structure model is first derived, then later refined to give an accurate model of the structure. The critical step is the deter- mination of the initial model. As materials problems decrease in length scale, the electron microscope has proven to be a versatile and effective tool for studying many problems. However, study of complex bulk structures by electron diffraction has been hindered by the problem of dynamical diffraction. This phenomenon makes bulk electron diffraction very sensitive to specimen thickness, and expensive equip- ment such as aberration-corrected scanning transmission microscopes or elaborate methodology such as high resolution imaging combined with diffraction and simulation are often required to generate good starting structures. The precession electron diffraction technique (PED), which has the ability to significantly reduce dynamical effects in diffraction patterns, has shown promise as being a “philosopher’s stone” for bulk electron diffraction. However, a comprehensive understanding of its abilities and limitations is necessary before it can be put into widespread use as a standalone technique. -

Direct Phase Determination in Protein Electron Crystallography
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 1791–1794, March 1997 Biophysics Direct phase determination in protein electron crystallography: The pseudo-atom approximation (electron diffractionycrystal structure analysisydirect methodsymembrane proteins) DOUGLAS L. DORSET Electron Diffraction Department, Hauptman–Woodward Medical Research Institute, Inc., 73 High Street, Buffalo, NY 14203-1196 Communicated by Herbert A. Hauptman, Hauptman–Woodward Medical Research Institute, Buffalo, NY, December 12, 1996 (received for review October 28, 1996) ABSTRACT The crystal structure of halorhodopsin is Another approach to such phasing problems, especially in determined directly in its centrosymmetric projection using cases where the structures have appropriate distributions of 6.0-Å-resolution electron diffraction intensities, without in- mass, would be to adopt a pseudo-atom approach. The concept cluding any previous phase information from the Fourier of using globular sub-units as quasi-atoms was discussed by transform of electron micrographs. The potential distribution David Harker in 1953, when he showed that an appropriate in the projection is assumed a priori to be an assembly of globular scattering factor could be used to normalize the globular densities. By an appropriate dimensional re-scaling, low-resolution diffraction intensities with higher accuracy than these ‘‘globs’’ are then assumed to be pseudo-atoms for the actual atomic scattering factors employed for small mol- normalization of the observed structure factors. After this ecule structures -

Solid State Physics: Problem Set #3 Structural Determination Via X-Ray Scattering Due: Friday Jan
Physics 375 Fall (12) 2003 Solid State Physics: Problem Set #3 Structural Determination via X-Ray Scattering Due: Friday Jan. 31 by 6 pm Reading assignment: for Monday, 3.2-3.3 (structure factors for different lattices) for Wednesday, 3.4-3.7 (scattering methods) for Friday, 4.1-4.3 (mechanical properties of crystals) Problem assignment: Chapter 3 Problems: *3.1 Debye-Scherrer analysis (identify the crystal structure) [Robert] 3.2 Lattice parameter from Debye-Scherrer data 3.11 Measuring thermal expansion with X-ray scattering A1. The unit cell dimension of fcc copper is 0.36 nm. Calculate the longest wavelength of X- rays which will produce diffraction from the close packed planes. From what planes could X- rays with l=0.50 nm be diffracted? *A2. Single crystal diffraction: A cubic crystal with lattice spacing 0.4 nm is mounted with its [001] axis perpendicular to an incident X-ray beam of wavelength 0.1 nm. Initially the crystal is set so as to produce a diffracted beam associated with the (020) planes. Calculate the angle through which the crystal must be turned in order to produce a beam from the (hkl) planes where: a) (hkl)=(030) b) (hkl)=(130) [Brian] c) Which of these diffracted beams would be forbidden if the crystal is: i) sc; ii) bcc; iii) fcc A3. Structure factors for the fcc and diamond bases. a) Construct the structure factor Shkl for the fcc lattice and show that it vanishes unless h, k, and l are all even or all odd. b) Construct the structure factor Shkl for the diamond lattice and show that it vanishes unless h, k, and l are all odd or h+k+l=4n where n is an integer. -
Simple Cubic Lattice
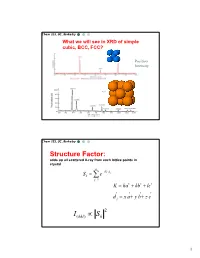
Chem 253, UC, Berkeley What we will see in XRD of simple cubic, BCC, FCC? Position Intensity Chem 253, UC, Berkeley Structure Factor: adds up all scattered X-ray from each lattice points in crystal n iKd j Sk e j1 K ha kb lc d j x a y b z c 2 I(hkl) Sk 1 Chem 253, UC, Berkeley X-ray scattered from each primitive cell interfere constructively when: eiKR 1 2d sin n For n-atom basis: sum up the X-ray scattered from the whole basis Chem 253, UC, Berkeley ' k d k d di R j ' K k k Phase difference: K (di d j ) The amplitude of the two rays differ: eiK(di d j ) 2 Chem 253, UC, Berkeley The amplitude of the rays scattered at d1, d2, d3…. are in the ratios : eiKd j The net ray scattered by the entire cell: n iKd j Sk e j1 2 I(hkl) Sk Chem 253, UC, Berkeley For simple cubic: (0,0,0) iK0 Sk e 1 3 Chem 253, UC, Berkeley For BCC: (0,0,0), (1/2, ½, ½)…. Two point basis 1 2 iK ( x y z ) iKd j iK0 2 Sk e e e j1 1 ei (hk l) 1 (1)hkl S=2, when h+k+l even S=0, when h+k+l odd, systematical absence Chem 253, UC, Berkeley For BCC: (0,0,0), (1/2, ½, ½)…. Two point basis S=2, when h+k+l even S=0, when h+k+l odd, systematical absence (100): destructive (200): constructive 4 Chem 253, UC, Berkeley Observable diffraction peaks h2 k 2 l 2 Ratio SC: 1,2,3,4,5,6,8,9,10,11,12. -

CHEM 3030 Introduction to X-Ray Crystallography X-Ray Diffraction Is the Premier Technique for the Determination of Molecular
CHEM 3030 Introduction to X-ray Crystallography X-ray diffraction is the premier technique for the determination of molecular structure in chemistry and biochemistry. There are three distinct parts to a structural determination once a high quality single crystal is grown and mounted on the diffractometer. 1. Geometric data collection – the unit cell dimensions are determined from the angles of a few dozen reflections. 2. Intensity Data Collection – the intensity of several thousand reflections are measured. 3. Structure solution – using Direct or Patterson methods, the phase problem is cracked and a function describing the e-density in the unit cell is generated (Fourier synthesis) from the measured intensities. Least squares refinement then optimizes agreement between Fobs (from Intensity data) and Fcalc (from structure). The more tedious aspects of crystallography have been largely automated and the computations are now within the reach of any PC. Crystallography provides an elegant application of symmetry concepts, mathematics (Fourier series), and computer methods to a scientific problem. Crystal structures are now ubiquitous in chemistry and biochemistry. This brief intro is intended to provide the minimum needed to appreciate literature data. FUNDAMENTALS 1. The 7 crystal systems and 14 Bravais lattices. 2. Space group Tables, special and general positions, translational symmetry elements, screw axes and glide planes. 3. Braggs law. Reflections occur only for integral values of the indices hkl because the distance traveled by an X-ray photon through a unit cell must coincide with an integral number of wavelengths. When this condition is satisfied scattering contributions from all unit cells add to give a net scattered wave with intensity I hkl at an angle Θhkl . -

Chapter 3 X-Ray Diffraction • Bragg's Law • Laue's Condition
Chapter 3 X-ray diffraction • Bragg’s law • Laue’s condition • Equivalence of Bragg’s law and Laue’s condition • Ewald construction • geometrical structure factor 1 Bragg’s law Consider a crystal as made out of parallel planes of ions, spaced a distance d apart. The conditions for a sharp peak in the intensity of the scattered radiation are 1. That the x-rays should be specularly reflected by the ions in any one plane and 2. That the reflected rays from successive planes should interfere constructively Path difference between two rays reflected from adjoining planes: 2d sin θ For the rays to interfere constructively, this path difference must be an integral number of wavelength λ nλ = 2d sin θ Bragg’s condition. 2 Bragg angle θ is just the half of the total angle 2 θ by which the incident beam is deflected. There are different ways of sectioning the crystal into planes, each of which will it self produce further reflection. The same portion of Bravais lattice shown in the previous page, with a different way of sectioning the crystal planes. The incident ray is the same. But both the direction and wavelength (determined by Bragg condition with d replaced by d’) of the reflected ray are different from the previous page. 3 Von Laue formulation of X-ray diffraction by a crystal • No particular sectioning of crystal planes r • Regard the crystal as composed of identical microscopic objects placed at Bravais lattice site R • Each of the object at lattice site reradiate the incident radiation in all directions. -

3.014 Materials Laboratory Oct. 13Th – Oct. 20Th , 2006 Lab Week 2 – Module Γ1 Derivative Structures Instructor: Meri Tr
3.014 Materials Laboratory Oct. 13th – Oct. 20th , 2006 Lab Week 2 – Module γ1 Derivative Structures Instructor: Meri Treska OBJECTIVES 9 Review principles of x-ray scattering from crystalline materials 9 Learn how to conduct x-ray powder diffraction experiments and use PDFs 9 Study the inter-relationship of different crystal structures SUMMARY OF TASKS 1) Calculate structure factor for materials to be investigated 2) Prepare samples for x-ray powder diffraction 2) Obtain x-ray scattering patterns for all materials 3) Compare obtained patterns with calculations and powder diffraction files (PDFs) 4) Perform peak fitting to determine percent crystallinity and crystallite size 1 BACKGROUND X-ray Diffraction from Crystalline Materials As discussed in 3.012, a periodic arrangement of atoms will give rise to constructive interference of scattered radiation having a wavelength λ comparable to the periodicity d when Bragg’s law is satisfied: nλ = 2sd inθ where n is an integer and θ is the angle of incidence. Bragg’s law tells us necessary conditions for diffraction, but provides no information regarding peak intensities. To use x-ray diffraction as a tool for materials identification, we must understand the relationship between structure/chemistry and the intensity of diffracted x-rays. Recall from 3.012 class that for a 1d array of atoms, the condition for constructive interference can be determined as follows: unit vector of ur diffracted beam S x ν a µ y ur S 0 x = a cos ν unit vector of ya= cos µ incident beam The total path difference: xy−=a cos ν − acos µλ= h uruurr (SS− 0 )⋅ a=hλ uruur r (SS− 0 ) Defining s = , the condition for 1d constructive interference becomes: λ r r s ⋅ ah= 2 r r For 3 dimensions, we have: s ⋅ ah= r r s ⋅bk= r r s ⋅ cl= where h, k and l are the Miller indices of the scattering plane. -

Polymers: Structure
Polymers: Structure W. Pyckhout-Hintzen This document has been published in Manuel Angst, Thomas Brückel, Dieter Richter, Reiner Zorn (Eds.): Scattering Methods for Condensed Matter Research: Towards Novel Applications at Future Sources Lecture Notes of the 43rd IFF Spring School 2012 Schriften des Forschungszentrums Jülich / Reihe Schlüsseltechnologien / Key Tech- nologies, Vol. 33 JCNS, PGI, ICS, IAS Forschungszentrum Jülich GmbH, JCNS, PGI, ICS, IAS, 2012 ISBN: 978-3-89336-759-7 All rights reserved. E 2 Polymers: Structure W. Pyckhout-Hintzen Jülich Centre for Neutron Science 1 & Institute for Complex Systems 1 Forschungszentrum Jülich GmbH Contents 1 Introduction............................................................................................... 2 2 Polymer Chain Models and Architecture............................................... 3 3 Scattering: intra- and inter-chain contributions.................................... 7 4 Blend of linear Polymers .......................................................................... 9 5 Tri-block copolymers..............................................................................13 6 Branched Chains .....................................................................................15 7 Summary..................................................................................................18 Appendix ............................................................................................................19 References ..........................................................................................................20 -

Section 2: X-Ray Diffraction and Reciprocal Lattice
Physics 927 E.Y.Tsymbal Section 2: X-ray Diffraction and Reciprocal Lattice Bragg law. Most methods for determining the atomic structure of crystals are based of the idea of scattering of radiation. X-rays is one of the types of the radiation which can be used. The wavelength of the radiation should have a wavelength comparable to a typical interatomic distance which is in solids of a few angstroms (10-8 cm). The x-ray wavelength λ can be estimated as follows hc 12.4 Eh==νλ⇒(Å) = . (2.1) λ Ek()eV Therefore, x-rays of energy 2-10 keV are suitable for studying the crystal structure. X-rays interact with electronic shells of atoms in a solid. Electrons absorb and re-radiate x-rays which can then be detected. Nuclei are too heavy to respond. The reflectivity of x-rays is of the order of 10-3- 10-5, so that the penetration in the solid is deep. Therefore, x-rays serve as a bulk probe. In 1913 Bragg found that crystalline solids have remarkably characteristic patterns of reflected x-ray radiation. In crystalline materials, for certain wavelengths and incident directions, intense peaks of scattered radiation were observed. Bragg accounted for this by regarding a crystal as made out of parallel planes of atoms, spaced by distance d apart. The conditions for a sharp peak in the intensity of the scattered radiation were that: (1) the x-rays should be specularly reflected by the atoms in one plane; (2) the reflected rays from the successive planes interfere constructively. -

Form and Structure Factors: Modeling and Interactions Jan Skov Pedersen, Inano Center and Department of Chemistry SAXS Lab University of Aarhus Denmark
Form and Structure Factors: Modeling and Interactions Jan Skov Pedersen, iNANO Center and Department of Chemistry SAXS lab University of Aarhus Denmark 1 New SAXS instrument in ultimo Nov 2014: Funding from: Research Council for Independent Research: Natural Science The Carlsberg Foundation The Novonordisk Foundation 2 Liquid metal jet X-ray sourcec Gallium Kα X-rays (λ = 1.34 Å) 3 x 109 photons/sec!!! 3 Special Optics Scatterless slits + optimized geometry: High Performance 2D 10 3 x 10 ph/sec!!! Detector Stopped-flow for rapid mixing Sample robot 4 Outline • Model fitting and least-squares methods • Available form factors ex: sphere, ellipsoid, cylinder, spherical subunits… ex: polymer chain • Monte Carlo integration for form factors of complex structures • Monte Carlo simulations for form factors of polymer models • Concentration effects and structure factors Zimm approach Spherical particles Elongated particles (approximations) Polymers 5 Motivation for ‘modelling’ - not to replace shape reconstruction and crystal-structure based modeling – we use the methods extensively - alternative approaches to reduce the number of degrees of freedom in SAS data structural analysis (might make you aware of the limited information content of your data !!!) - provide polymer-theory based modeling of flexible chains - describe and correct for concentration effects 6 Literature Jan Skov Pedersen, Analysis of Small-Angle Scattering Data from Colloids and Polymer Solutions: Modeling and Least-squares Fitting (1997). Adv. Colloid Interface Sci., 70, 171-210. Jan Skov Pedersen Monte Carlo Simulation Techniques Applied in the Analysis of Small-Angle Scattering Data from Colloids and Polymer Systems in Neutrons, X-Rays and Light P. Lindner and Th.