Strategies to Obtain Diverse and Specific Human Monoclonal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Precision Medicine for Human Cancers with Notch Signaling Dysregulation (Review)
INTERNATIONAL JOURNAL OF MOleCular meDICine 45: 279-297, 2020 Precision medicine for human cancers with Notch signaling dysregulation (Review) MASUKO KATOH1 and MASARU KATOH2 1M & M PrecMed, Tokyo 113-0033; 2Department of Omics Network, National Cancer Center, Tokyo 104-0045, Japan Received September 16, 2019; Accepted November 20, 2019 DOI: 10.3892/ijmm.2019.4418 Abstract. NOTCH1, NOTCH2, NOTCH3 and NOTCH4 are conjugate (ADC) Rova-T, and DLL3-targeting chimeric antigen transmembrane receptors that transduce juxtacrine signals of receptor‑modified T cells (CAR‑Ts), AMG 119, are promising the delta-like canonical Notch ligand (DLL)1, DLL3, DLL4, anti-cancer therapeutics, as are other ADCs or CAR-Ts targeting jagged canonical Notch ligand (JAG)1 and JAG2. Canonical tumor necrosis factor receptor superfamily member 17, Notch signaling activates the transcription of BMI1 proto-onco- CD19, CD22, CD30, CD79B, CD205, Claudin 18.2, fibro- gene polycomb ring finger, cyclin D1, CD44, cyclin dependent blast growth factor receptor (FGFR)2, FGFR3, receptor-type kinase inhibitor 1A, hes family bHLH transcription factor 1, tyrosine-protein kinase FLT3, HER2, hepatocyte growth factor hes related family bHLH transcription factor with YRPW receptor, NECTIN4, inactive tyrosine-protein kinase 7, inac- motif 1, MYC, NOTCH3, RE1 silencing transcription factor and tive tyrosine-protein kinase transmembrane receptor ROR1 transcription factor 7 in a cellular context-dependent manner, and tumor-associated calcium signal transducer 2. ADCs and while non-canonical Notch signaling activates NF-κB and Rac CAR-Ts could alter the therapeutic framework for refractory family small GTPase 1. Notch signaling is aberrantly activated cancers, especially diffuse-type gastric cancer, ovarian cancer in breast cancer, non-small-cell lung cancer and hematological and pancreatic cancer with peritoneal dissemination. -

Minutes of the CHMP Meeting 14-17 September 2020
13 January 2021 EMA/CHMP/625456/2020 Corr.1 Human Medicines Division Committee for medicinal products for human use (CHMP) Minutes for the meeting on 14-17 September 2020 Chair: Harald Enzmann – Vice-Chair: Bruno Sepodes Disclaimers Some of the information contained in these minutes is considered commercially confidential or sensitive and therefore not disclosed. With regard to intended therapeutic indications or procedure scopes listed against products, it must be noted that these may not reflect the full wording proposed by applicants and may also vary during the course of the review. Additional details on some of these procedures will be published in the CHMP meeting highlights once the procedures are finalised and start of referrals will also be available. Of note, these minutes are a working document primarily designed for CHMP members and the work the Committee undertakes. Note on access to documents Some documents mentioned in the minutes cannot be released at present following a request for access to documents within the framework of Regulation (EC) No 1049/2001 as they are subject to on- going procedures for which a final decision has not yet been adopted. They will become public when adopted or considered public according to the principles stated in the Agency policy on access to documents (EMA/127362/2006). 1 Addition of the list of participants Official address Domenico Scarlattilaan 6 ● 1083 HS Amsterdam ● The Netherlands Address for visits and deliveries Refer to www.ema.europa.eu/how-to-find-us Send us a question Go to www.ema.europa.eu/contact Telephone +31 (0)88 781 6000 An agency of the European Union © European Medicines Agency, 2020. -

Inflammation and Cardiometabolic Risk
Official Publication of the National Lipid Association LipidSpin Inflammation and Cardiometabolic Risk Also in this issue: Clinical Trial Evidence for the Role of Reducing Inflammation for Cardiovascular Disease Prevention Identifying and Managing the Patient with High Inflammatory Burden Dietary Patterns and Systemic Inflammation: Examining the Connection This issue is sponsored by the Pacific Lipid Association. Volume 17 Issue 1 2019 visit www.lipid.org NATIONAL LIPID ASSOCIATION Turnberry Isle Miami 19999 W. Country Club Dr. SCIENTIFIC 9 Aventura, FL 33180 1 SESSIONS Group Rate: $229/night++ 0 Reservation Cut-Off Date: MIAMI 2 April 14, 2019 Pre-Conference Courses | May 15-16 Make your plans to join the NLA in 2019 for these premier events! lipid.org/conferences FEBRUARY 22-24 SEPTEMBER 13-15 2019 2019 PORTLAND MINNEAPOLIS Hilton Portland Downtown Renaissance Minneapolis Hotel, the Depot 921 SW Sixth Avenue, Portland, OR 97204 225 Third Avenue South, Minneapolis, MN 55401 Group Rate: $185/night++ Group Rate: $179/night++ Reservation Cut-Off Date: January 21, 2019 Reservation Cut-Off Date: August 14, 2019 Pre-Conference Courses | February 21-22, 2019 Pre-Conference Courses | September 12-13, 2019 In This Issue: 2019 (Volume 17, Issue 1) Editors DANIEL E. SOFFER, MD, FNLA* 2 From the NLA President 18 Practical Pearls Clinical Associate Professor of Medicine New Clinical Guidelines: Paving the Top 10 Ways to Reduce Inflammation University of Pennsylvania — Cezary Wójcik, MD, PhD, DSc, FNLA* Internal Medicine and Preventive Cardiology Road Ahead University of Pennsylvania Health System — Alan S. Brown, MD, FNLA* Philadelphia, PA — Daniel E. Soffer MD, FNLA* — Kaye-Eileen Willard MD, FNLA* KAYE-EILEEN WILLARD, MD, FNLA* 20 Case Study Medical Director, Lipid Clinic and Physician Advisor Identifying and Managing the Patient Ascension Wisconsin All Saints Racine, WI 4 From the PLA President with High Inflammatory Burden President’s Views — Michelle Taylor, CCRN, ANP Associate Editors — Nathan D. -

Predictive QSAR Tools to Aid in Early Process Development of Monoclonal Antibodies
Predictive QSAR tools to aid in early process development of monoclonal antibodies John Micael Andreas Karlberg Published work submitted to Newcastle University for the degree of Doctor of Philosophy in the School of Engineering November 2019 Abstract Monoclonal antibodies (mAbs) have become one of the fastest growing markets for diagnostic and therapeutic treatments over the last 30 years with a global sales revenue around $89 billion reported in 2017. A popular framework widely used in pharmaceutical industries for designing manufacturing processes for mAbs is Quality by Design (QbD) due to providing a structured and systematic approach in investigation and screening process parameters that might influence the product quality. However, due to the large number of product quality attributes (CQAs) and process parameters that exist in an mAb process platform, extensive investigation is needed to characterise their impact on the product quality which makes the process development costly and time consuming. There is thus an urgent need for methods and tools that can be used for early risk-based selection of critical product properties and process factors to reduce the number of potential factors that have to be investigated, thereby aiding in speeding up the process development and reduce costs. In this study, a framework for predictive model development based on Quantitative Structure- Activity Relationship (QSAR) modelling was developed to link structural features and properties of mAbs to Hydrophobic Interaction Chromatography (HIC) retention times and expressed mAb yield from HEK cells. Model development was based on a structured approach for incremental model refinement and evaluation that aided in increasing model performance until becoming acceptable in accordance to the OECD guidelines for QSAR models. -
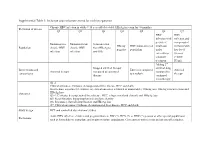
Inclusion and Exclusion Criteria for Each Key Question
Supplemental Table 1: Inclusion and exclusion criteria for each key question Chronic HBV infection in adults ≥ 18 year old (detectable HBsAg in serum for >6 months) Definition of disease Q1 Q2 Q3 Q4 Q5 Q6 Q7 HBV HBV infection with infection and persistent compensated Immunoactive Immunotolerant Seroconverted HBeAg HBV mono-infected viral load cirrhosis with Population chronic HBV chronic HBV from HBeAg to negative population under low level infection infection anti-HBe entecavir or viremia tenofovir (<2000 treatment IU/ml) Adding 2nd Stopped antiviral therapy antiviral drug Interventions and Entecavir compared Antiviral Antiviral therapy compared to continued compared to comparisons to tenofovir therapy therapy continued monotherapy Q1-2: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Intermediate outcomes (if evidence on clinical outcomes is limited or unavailable): HBsAg loss, HBeAg seroconversion and Outcomes HBeAg loss Q3-4: Cirrhosis, decompensated liver disease, HCC, relapse (viral and clinical) and HBsAg loss Q5: Renal function, hypophosphatemia and bone density Q6: Resistance, flare/decompensation and HBeAg loss Q7: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Study design RCT and controlled observational studies Acute HBV infection, children and pregnant women, HIV (+), HCV (+) or HDV (+) persons or other special populations Exclusions such as hemodialysis, transplant, and treatment failure populations. Co treatment with steroids and uncontrolled studies. Supplemental Table 2: Detailed Search Strategy: Ovid Database(s): Embase 1988 to 2014 Week 37, Ovid MEDLINE(R) In-Process & Other Non- Indexed Citations and Ovid MEDLINE(R) 1946 to Present, EBM Reviews - Cochrane Central Register of Controlled Trials August 2014, EBM Reviews - Cochrane Database of Systematic Reviews 2005 to July 2014 Search Strategy: # Searches Results 1 exp Hepatitis B/dt 26410 ("hepatitis B" or "serum hepatitis" or "hippie hepatitis" or "injection hepatitis" or 2 178548 "hepatitis type B").mp. -

(12) United States Patent (10) Patent No.: US 9.468,689 B2 Zeng Et Al
USOO9468689B2 (12) United States Patent (10) Patent No.: US 9.468,689 B2 Zeng et al. (45) Date of Patent: *Oct. 18, 2016 (54) ULTRAFILTRATION CONCENTRATION OF (56) References Cited ALLOTYPE SELECTED ANTIBODES FOR SMALL-VOLUME ADMINISTRATION U.S. PATENT DOCUMENTS 5,429,746 A 7/1995 Shadle et al. (71) Applicant: Immunomedics, Inc., Morris Plains, NJ 5,789,554 A 8/1998 Leung et al. (US) 6,171,586 B1 1/2001 Lam et al. 6,187,287 B1 2/2001 Leung et al. (72) Inventors: Li Zeng, Edison, NJ (US); Rohini 6,252,055 B1 6/2001 Relton Mitra, Brigdewater, NJ (US); Edmund 6,676,924 B2 1/2004 Hansen et al. 6,870,034 B2 3/2005 Breece et al. A. Rossi, Woodland Park, NJ (US); 6,893,639 B2 5/2005 Levy et al. Hans J. Hansen, Picayune, MS (US); 6,991,790 B1 1/2006 Lam et al. David M. Goldenberg, Mendham, NJ 7,038,017 B2 5, 2006 Rinderknecht et al. (US) 7,074,403 B1 7/2006 Goldenberg et al. 7,109,304 B2 9, 2006 Hansen et al. 7,138,496 B2 11/2006 Hua et al. (73) Assignee: Immunomedics, Inc., Morris Plains, NJ 7,151,164 B2 * 12/2006 Hansen et al. ............. 530,387.3 (US) 7,238,785 B2 7/2007 Govindan et al. 7,251,164 B2 7/2007 Okhonin et al. (*) Notice: Subject to any disclaimer, the term of this 7.282,567 B2 10/2007 Goldenberg et al. patent is extended or adjusted under 35 7,300,655 B2 11/2007 Hansen et al. -

Activating Death Receptor DR5 As a Therapeutic Strategy for Rhabdomyosarcoma
International Scholarly Research Network ISRN Oncology Volume 2012, Article ID 395952, 10 pages doi:10.5402/2012/395952 Review Article Activating Death Receptor DR5 as a Therapeutic Strategy for Rhabdomyosarcoma Zhigang Kang,1, 2 Shi-Yong Sun,3 and Liang Cao1 1 Genetics Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD 20892, USA 2 Laboratory of Proteomics and Analytical Technologies, SAIC-Frederick, Inc., NCI Frederick, Frederick, MD 21702, USA 3 Department of Hematology and Medical Oncology, Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA Correspondence should be addressed to Liang Cao, [email protected] Received 4 January 2012; Accepted 24 January 2012 Academic Editors: E. Boven and S. Mandruzzato Copyright © 2012 Zhigang Kang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children. It is believed to arise from skeletal muscle progenitors, preserving the expression of genes critical for embryonic myogenic development such as MYOD1 and myogenin. RMS is classified as embryonal, which is more common in younger children, or alveolar, which is more prevalent in elder children and adults. Despite aggressive management including surgery, radiation, and chemotherapy, the outcome for children with metastatic RMS is dismal, and the prognosis has remained unchanged for decades. Apoptosis is a highly regulated process critical for embryonic development and tissue and organ homeostasis. Like other types of cancers, RMS develops by evading intrinsic apoptosis via mutations in the p53 tumor suppressor gene. -

CNTO 888) Concentration Time Data
Pharmacokinetics and Pharmacodynamics The Journal of Clinical Pharmacology Utilizing Pharmacokinetics/Pharmacodynamics 53(10) 1020–1027 ©2013, The American College of Modeling to Simultaneously Examine Clinical Pharmacology DOI: 10.1002/jcph.140 Free CCL2, Total CCL2 and Carlumab (CNTO 888) Concentration Time Data Gerald J. Fetterly, PhD1, Urvi Aras, PhD1, Patricia D. Meholick, MS1, Chris Takimoto, MD, PhD2, Shobha Seetharam, PhD2, Thomas McIntosh, MS2, Johann S. de Bono, MD, PhD3, Shahneen K. Sandhu, MD3, Anthony Tolcher, MD4, Hugh M. Davis, PhD2, Honghui Zhou, PhD, FCP2, and Thomas A. Puchalski, PharmD2 Abstract The chemokine ligand 2 (CCL2) promotes angiogenesis, tumor proliferation, migration, and metastasis. Carlumab is a human IgG1k monoclonal antibody with high CCL2 binding affinity. Pharmacokinetic/pharmacodynamic data from 21 cancer patients with refractory tumors were analyzed. The PK/PD model characterized the temporal relationships between serum concentrations of carlumab, free CCL2, and the carlumab–CCL2 complex. Dose‐dependent increases in total CCL2 concentrations were observed and were consistent with shifting free CCL2. Free CCL2 declined rapidly after the initial carlumab infusion, returned to baseline within 7 days, and increased to levels greater than baseline following subsequent doses. Mean predicted half‐lives of carlumab and carlumab–CCL2 complex were approximately 2.4 days and approximately 1 hour for free CCL2. The mean dissociation constant (KD), 2.4 nM, was substantially higher than predicted by in vitro experiments, -

Form 10-Q Regeneron Pharmaceuticals, Inc
UNITED STATES SECURITIES AND EXCHANGE COMMISSION Washington, DC 20549 FORM 10-Q (Mark One) QUARTERLY REPORT PURSUANT TO SECTION 13 OR 15(d) OF THE SECURITIES (X) EXCHANGE ACT OF 1934 For the quarterly period ended March 31, 2013 OR TRANSITION REPORT PURSUANT TO SECTION 13 OR 15(d) OF THE SECURITIES ( ) EXCHANGE ACT OF 1934 For the transition period from __________ to __________ Commission File Number 0-19034 REGENERON PHARMACEUTICALS, INC. (Exact name of registrant as specified in its charter) New York 13-3444607 (State or other jurisdiction of (I.R.S. Employer Identification No.) incorporation or organization) 777 Old Saw Mill River Road, Tarrytown, New York 10591-6707 (Address of principal executive offices) (Zip Code) (914) 847-7000 (Registrant’s telephone number, including area code) Indicate by check mark whether the registrant: (1) has filed all reports required to be filed by Section 13 or 15(d) of the Securities Exchange Act of 1934 during the preceding 12 months (or for such shorter period that the registrant was required to file such reports), and (2) has been subject to such filing requirements for the past 90 days. Yes X No Indicate by check mark whether the registrant has submitted electronically and posted on its corporate Web site, if any, every Interactive Data File required to be submitted and posted pursuant to Rule 405 of Regulation S-T (§232.405 of this chapter) during the preceding 12 months (or for such shorter period that the registrant was required to submit and post such files). Yes X No Indicate by check mark whether the registrant is a large accelerated filer, an accelerated filer, a non-accelerated filer, or a smaller reporting company. -

WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C07D 519/00 (2006.01) A61P 39/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C07D 487/04 (2006.01) A61P 35/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 31/5517 (2006.01) A61P 37/00 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, A61K 47/48 (2006.01) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (21) International Application Number: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, PCT/IB2013/058229 SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (22) International Filing Date: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2 September 2013 (02.09.2013) ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: HANGZHOU DAC BIOTECH CO., LTD UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, [US/CN]; Room B2001-B2019, Building 2, No 452 Sixth TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, Street, Hangzhou Economy Development Area, Hangzhou EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, City, Zhejiang 310018 (CN). -

Classification Decisions Taken by the Harmonized System Committee from the 47Th to 60Th Sessions (2011
CLASSIFICATION DECISIONS TAKEN BY THE HARMONIZED SYSTEM COMMITTEE FROM THE 47TH TO 60TH SESSIONS (2011 - 2018) WORLD CUSTOMS ORGANIZATION Rue du Marché 30 B-1210 Brussels Belgium November 2011 Copyright © 2011 World Customs Organization. All rights reserved. Requests and inquiries concerning translation, reproduction and adaptation rights should be addressed to [email protected]. D/2011/0448/25 The following list contains the classification decisions (other than those subject to a reservation) taken by the Harmonized System Committee ( 47th Session – March 2011) on specific products, together with their related Harmonized System code numbers and, in certain cases, the classification rationale. Advice Parties seeking to import or export merchandise covered by a decision are advised to verify the implementation of the decision by the importing or exporting country, as the case may be. HS codes Classification No Product description Classification considered rationale 1. Preparation, in the form of a powder, consisting of 92 % sugar, 6 % 2106.90 GRIs 1 and 6 black currant powder, anticaking agent, citric acid and black currant flavouring, put up for retail sale in 32-gram sachets, intended to be consumed as a beverage after mixing with hot water. 2. Vanutide cridificar (INN List 100). 3002.20 3. Certain INN products. Chapters 28, 29 (See “INN List 101” at the end of this publication.) and 30 4. Certain INN products. Chapters 13, 29 (See “INN List 102” at the end of this publication.) and 30 5. Certain INN products. Chapters 28, 29, (See “INN List 103” at the end of this publication.) 30, 35 and 39 6. Re-classification of INN products. -

Tanibirumab (CUI C3490677) Add to Cart
5/17/2018 NCI Metathesaurus Contains Exact Match Begins With Name Code Property Relationship Source ALL Advanced Search NCIm Version: 201706 Version 2.8 (using LexEVS 6.5) Home | NCIt Hierarchy | Sources | Help Suggest changes to this concept Tanibirumab (CUI C3490677) Add to Cart Table of Contents Terms & Properties Synonym Details Relationships By Source Terms & Properties Concept Unique Identifier (CUI): C3490677 NCI Thesaurus Code: C102877 (see NCI Thesaurus info) Semantic Type: Immunologic Factor Semantic Type: Amino Acid, Peptide, or Protein Semantic Type: Pharmacologic Substance NCIt Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor tyrosine kinase expressed by endothelial cells, while VEGF is overexpressed in many tumors and is correlated to tumor progression. PDQ Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor