Genetic Parameters Estimation and Breeding Values Prediction for Linear Described Traits in the Old Kladruber Horse*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

List of Horse Breeds 1 List of Horse Breeds
List of horse breeds 1 List of horse breeds This page is a list of horse and pony breeds, and also includes terms used to describe types of horse that are not breeds but are commonly mistaken for breeds. While there is no scientifically accepted definition of the term "breed,"[1] a breed is defined generally as having distinct true-breeding characteristics over a number of generations; its members may be called "purebred". In most cases, bloodlines of horse breeds are recorded with a breed registry. However, in horses, the concept is somewhat flexible, as open stud books are created for developing horse breeds that are not yet fully true-breeding. Registries also are considered the authority as to whether a given breed is listed as Light or saddle horse breeds a "horse" or a "pony". There are also a number of "color breed", sport horse, and gaited horse registries for horses with various phenotypes or other traits, which admit any animal fitting a given set of physical characteristics, even if there is little or no evidence of the trait being a true-breeding characteristic. Other recording entities or specialty organizations may recognize horses from multiple breeds, thus, for the purposes of this article, such animals are classified as a "type" rather than a "breed". The breeds and types listed here are those that already have a Wikipedia article. For a more extensive list, see the List of all horse breeds in DAD-IS. Heavy or draft horse breeds For additional information, see horse breed, horse breeding and the individual articles listed below. -

History-Of-Breeding-And-Training-Of-The-Kladruber-Horses
History of Breeding and Training of the Kladruber Horses The Kladruber horse is the only breed of the original ceremonial horses still bred that is the only draught horse breed in the world originated, bred and trained for drawing carriages of the social elites. Thanks to the Habsburg conservatism and unchanged breeding goal, the Kladruber horse has preserved its original “baroque” appearance from the 18th century to date. It still bears the traits of the original, but now extinct breeds (old Spanish horse and old Italian horse) which were at its beginning and from medieval times until the 18th century influenced the stock in most European countries and colonies and by the end of the 18th century were extinct. Even though there are only limited opportunities for ceremonial carriage horses to be used at (now the most frequent breeds are warmblooded horses for sport) the Kladruber horse breed has been preserved and still serves its original purpose for example at the Danish Royal Court and it is also used for state functions. Horse breeds are divided into primitive (indigenous) and intentionally designed (on the basis of targeted selective breeding) however some breeds oscillate between these two main types. Then the horse breeds are divided according to their purpose such as draught horses which the carriage horses fall into (weight up to 1200 kg), riding horses (up to 800kg) and pack horses (less than 500 kg). A new horse breed came into existence either in a particular area, using the same genetic material and the effect of the external conditions and climate (most of the breeds started in this way) or it came into existence in a single place – at a dedicated stud farm with a clearly defined breeding goal using particular horses of selected breeds imported for this sole purpose and applying the knowledge of selective breeding available at that time as well as the knowledge of local natural conditions and climate. -

Electronic Supplementary Material - Appendices
1 Electronic Supplementary Material - Appendices 2 Appendix 1. Full breed list, listed alphabetically. Breeds searched (* denotes those identified with inherited disorders) # Breed # Breed # Breed # Breed 1 Ab Abyssinian 31 BF Black Forest 61 Dul Dülmen Pony 91 HP Highland Pony* 2 Ak Akhal Teke 32 Boe Boer 62 DD Dutch Draft 92 Hok Hokkaido 3 Al Albanian 33 Bre Breton* 63 DW Dutch Warmblood 93 Hol Holsteiner* 4 Alt Altai 34 Buc Buckskin 64 EB East Bulgarian 94 Huc Hucul 5 ACD American Cream Draft 35 Bud Budyonny 65 Egy Egyptian 95 HW Hungarian Warmblood 6 ACW American Creme and White 36 By Byelorussian Harness 66 EP Eriskay Pony 96 Ice Icelandic* 7 AWP American Walking Pony 37 Cam Camargue* 67 EN Estonian Native 97 Io Iomud 8 And Andalusian* 38 Camp Campolina 68 ExP Exmoor Pony 98 ID Irish Draught 9 Anv Andravida 39 Can Canadian 69 Fae Faeroes Pony 99 Jin Jinzhou 10 A-K Anglo-Kabarda 40 Car Carthusian 70 Fa Falabella* 100 Jut Jutland 11 Ap Appaloosa* 41 Cas Caspian 71 FP Fell Pony* 101 Kab Kabarda 12 Arp Araappaloosa 42 Cay Cayuse 72 Fin Finnhorse* 102 Kar Karabair 13 A Arabian / Arab* 43 Ch Cheju 73 Fl Fleuve 103 Kara Karabakh 14 Ard Ardennes 44 CC Chilean Corralero 74 Fo Fouta 104 Kaz Kazakh 15 AC Argentine Criollo 45 CP Chincoteague Pony 75 Fr Frederiksborg 105 KPB Kerry Bog Pony 16 Ast Asturian 46 CB Cleveland Bay 76 Fb Freiberger* 106 KM Kiger Mustang 17 AB Australian Brumby 47 Cly Clydesdale* 77 FS French Saddlebred 107 KP Kirdi Pony 18 ASH Australian Stock Horse 48 CN Cob Normand* 78 FT French Trotter 108 KF Kisber Felver 19 Az Azteca -

Multiple Pregnancy in Mares
MENDELNET 2016 MULTIPLE PREGNANCY IN MARES MARIE IMRICHOVA Department of Animal Breeding Mendel University in Brno Zemedelska 1, 613 00 Brno CZECH REPUBLIC [email protected] Abstract: Mare is in terms of reproduction described as uniparous, it means that she has one foal. More embryos, fetuses or foals represents non – physiological phenomenon and as such it brings a lot of complications. In terms of the etiology is discussed as a predisposing factor mare’s breed, the most common is a higher incidence associated with Thoroughbreds. Although in context of multiple pregnancies often mentioned is mare‘s age. The aim of this study was to evaluate the incidence and results of multiple pregnancy in Thoroughbred and Old Kladruber Horse mares. It was found 323 records of multiple pregnancies in Thoroughbred and 48 in Old Kladruber Horse mares and the result of the multiple pregnancy was in Thoroughbreds in 258 cases twin abortion, in 45 cases parturition of 2 dead foals, 15 records of parturition 1 live and 1 dead foal and in 5 cases it was the parturition of 2 live foals. In Old Kladruber Horse twin abortion was recorded 36 times, in 7 cases 2 dead foals and in 5 cases 2 live foals. Key Words: horse, breeding, reproduction, pregnancy, twins INTRODUCTION The reproduction standard in horse breeding is the fundamental informative factor affecting its success and profitability. One of the factors that can decide about the reproduction outcome is also multiple pregnancy. Multiple pregnancy basically may occur naturally either by spontaneous division of embryos, or by fertilization of two oocytes after multiple ovulation. -

Lipizzan Laurels United States Lipizzan Federation©
Lipizzan Laurels United States Lipizzan Federation© Awards Program The United States Lipizzan Federation (USLF) recognizes Lipizzans/XL Lipizzans competing in events against all breeds with two special award programs—the USLF Lipizzan/XL Lipizzan Laurels Award and the USLF Lipizzan/XL Lipizzan Star Award. Award participants automatically are entered in both programs when they submit results. The USLF Lipizzan Laurels presents a Lipizzan Laurels Award for outstanding performance by a Lipizzan/XL Lipizzan horse in several sections within the program’s ten major disciplines. Junior Exhibitor Awards are also presented in all ten disciplines. Lipizzan Laurels awards are tabulated on an annual basis during the competition year, which runs from November 1st of the previous year to October 31st of the current year. The Lipizzan/XL Lipizzan Star Award is a lifetime award presented to each Lipizzan/XL Lipizzan meeting Star requirements. Lipizzan/XL Lipizzan horses may take as many years as needed to earn Star points. The owner of the achieving horse will receive an official bronze, silver, gold, or platinum Star when they have accumulated the necessary points in their discipline. Stars are presented in ten disciplines: Show, Competitive Trail, Endurance, Dressage, Eventing, Working Western, Carriage Pleasure, Combined Driving, Western Dressage, and Working Equitation. Horses who earn five of the ten bronze Stars are presented with the USLF Lipizzan/XL Lipizzan Sport Horse Award. 1 GENERAL GUIDELINES To be eligible for these awards, horses must be registered with the USLF at the time scores are earned. Both the owner and all riders or drivers of the horse must be current USLF members at the time scores are earned. -

Quantitative Genetic Analysis of Melanoma and Grey Level in Lipizzan Horses
7th World Congress on Genetics Applied to Livestock Production, August 19-23, 2002, Montpellier, France QUANTITATIVE GENETIC ANALYSIS OF MELANOMA AND GREY LEVEL IN LIPIZZAN HORSES I. Curik1, M. Seltenhammer2 and J. Sölkner3 1Animal Science Department, Faculty of Agriculture, University of Zagreb, Croatia 2Clinic of Surgery and Ophthalmology, University of Veterinary Medicine Vienna, Austria 3Department of Livestock Science, University of Agricultural Sciences Vienna, Austria INTRODUCTION Changes of coat colour in which the "dark (non-grey)" colour present in foals is progressively replaced by grey is a known phenomenon in horses. A similar process is present in humans. The grey coat colour is inherited as a dominant trait and is the characteristic, although not exclusive, colour for some horse breeds (Bowling, 2000). The Lipizzan horse, originally bred for show and parade at the Imperial Court in Vienna, is among those breeds. Unfortunately, melanomas (skin tumours) are more prevalent in grey than in non-grey horses (e.g. Seltenhammer, 2000). The causative relationship for this positive association as well as the molecular basis for both traits (melanoma and grey level) are not known (Rieder, 1999 ; Seltenhammer, 2000). The inheritance of coat colour in horses has been always studied from a qualitative view (Sponenberg, 1996 ; Bowling, 2000). In the present study we quantified the grey level (shade) and estimated the proportion of additive genetic component (heritability in the narrow sense) of this trait. Further, we estimated the genetic relationship between melanoma stages and grey level as well as the additive inheritance of melanoma stages. MATERIAL AND METHODS Horses. Data for this study was collected from 351 grey Lipizzan horses of four national studs (Djakovo – Croatia ; 64 horses, Piber – Austria ; 160 horses, Szilvesvarad – Hungary ; 67 horses and Topol'cianky – Slovakia ; 60 horses). -

History-And-Development
History and Development The oldest history The Landscape offers more than 500 years of experience of how the people who bred horses in individual historical periods considered the sustainable use of landscape, and of natural resources and their cultivation. The terrain, soil, climate, water and vegetation were used and also cultivated to the extent the then farming customs and degree of knowledge allowed for breeding horses for an important European Imperial Court. The landscape still bears the authentic traces of this development since the oldest days. In the same way the horse breeding and training know-how that became the main source of income for local population was developing. Hand in hand with this development these people were also taking care of the landscape. With the time the “mere” farming cultivation of natural countryside turned into the work of architecture while the art plan did not overrule the farming purpose. The aesthetics were always considered the natural quality of work. Since horses and particularly coach and carriage horses owned by the Emperor evoke noble feelings in men, the Landscape is naturally also refined. Numerous archive and historical documents bear evidence of centuries’ long continuous and rich development of architectural heritage in the Landscape. The buildings that were raised, pulled down and refurbished there in the course of times complement the landscape and they are only a part of its memory and current appearance. The history of horse breeding in Kladruby is probably as old as the history of the local settlement. The Elbe river alluvial plain, the location of the Landscape, had been settled since medieval times40, later than the neighbouring fertile flatlands further away from the river. -

Vol. 17, Issue 4.Indd
HAUTE ÉCOLE Quarterly Publication of the Lipizzan Association of North America Volume 17, Issue 4 The Naming System Within the Lipizzan Race Approved & Unapproved Breeding Practices Registry Office Open for Business 2009 Summer Clinics Cloning The naming system within the Lipizzan race . Traditional and practical By Atjan Hop Everybody who is interested in the back- grounds of the breeding and breeding histo- ry of the Lipizzaner horse breed will sooner or later be faced with the traditional beauti- ful, but repeating naming system. At first sight the names sound elegant, completely suitable with this noble imperial horse breed. But in daily practice the system ap- pears to be confusing, with all those simi- lar names and roman numerals. And what about the different system, that is being used in Hungary and Romania? Let’s try to explain the principals and backgrounds. Atjan Hop & Neapolitano Elvira working on Long Reins In the Renaissance and Baroque time era, the Spanish horse type was the leading type among the European nobility. The court habits of the prominent noble dynasty since the 16th century, the Habsburgs, were taken over by all other nobility in Europe, including the dominant use of the mag- nificent horse breed of the Iberian Peninsula. These proud, elegant and brave horses with their elevated movements really did fit the demands of the European nobility in those times. Those horses were not only directly purchased on the Iberian Peninsula, but also bred in a number of court studfarms throughout the whole of Europe, in order to fulfil the demand directly. The Habsburg dynasty kept their leading role in providing these horses. -
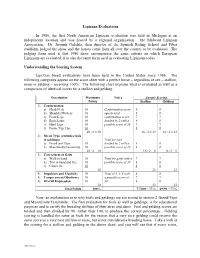
2-Evaluation Summary (H0933573
Lipizzan Evaluations In 1986, the first North American Lipizzan evaluation was held in Michigan at an independent location and was hosted by a regional organization – the Midwest Lipizzan Association. Dr. Jaromir Oulehla, then director of the Spanish Riding School and Piber studfarm, judged the show and the horses came from all over the country to be evaluated. The judging form used at that 1986 show encompasses the same criteria on which European Lipizzans are evaluated; it is also the same form used in evaluating Lipizzans today. Understanding the Scoring System Lipizzan breed evaluations have been held in the United States since 1986. The following categories appear on the score sheet with a perfect horse – regardless of sex – stallion, mare or gelding – receiving 100%. The following chart explains what is evaluated as well as a comparison of identical scores for a stallion and gelding. Description Maximum Notes Sample Scoring Points Stallion Gelding 1) Conformation a) Head/Neck 10 Conformation score 8 8 b) Shoulder/Withers 10 equals total 8 8 c) Front Legs 10 confirmation score 7 7 d) Back/Loins 10 divided by 2 with a 8 8 e) Hind Legs 10 possible score of 30 7 7 f) Frame/Top Line 10 8 8 60 / 2 = 30 46 / 2 = 23 46 / 2 = 23 2) Breed Type (excludes foals & geldings) Total for type a) Breed and Type 10 divided by 2 with a 8 0 b) Masculinity/Femininity 10 possible score of 10 8 0 20 / 2 = 10 16 / 2 = 8 0 / 2 = 0 3) Correctness of Gaits a) Walk in hand 10 Total for gaits with a 7 7 b) Trot in hand and free 10 possible score of 30 8 8 c) Canter free 10 8 8 30 23 23 4) Impulsion and Elasticity 10 Total of 4, 5, 6 with 8 8 5) Temperament/Obedience 10 a possible score of 7 7 6) Overall Impression 10 30 8 8 30 23 23 Total Points 100% 77/100 = 77% 69/90 = 77% Now, an explanation as to why foals and geldings are not scored in section 2 Breed/Type and Masculinity/Femininity. -

20091112 Organismi Passaporto Equidi
Ministero delle politiche agricole alimentari e forestali Dipartimento delle Politiche competitive del mondo rurale e della qualità Ex Direzione Generale dello Sviluppo Rurale, delle Infrastrutture e dei Servizi Ufficio SVIRIS X - Produzioni Animali - Dirigente: Francesco Scala Tel. 06 46655098-46655096 - 06 484459 Fax. 06 46655132 e-mail: [email protected] web: www.politicheagricole.it ELENCO ORGANISMI EMITTENTI PASSAPORTO EQUIDI REG. (CE) N. 504/2008 ART. 4 COMMA 5 LIST OF AGENCIES RELEASING EQUINE PASSPORT REG. (CE) N.504/2008 ART.4, COMMA 5 Codice UELN Libro genealogico/ Razze Contatti Associazione/Organizzazione UELN code Registro anagrafico Races Contacts Associations/Organizations Stud book/population register AIA-Associazione Italiana 380001 Registro Anagrafico Equine: Dr. Giancarlo Allevatori razze equine ed Cavallino della Giara Carchedi Via Tomassetti, 9 asinine a limitata Cavallino di Monterufoli 00161 ROMA diffusione Cavallo del Catria Cavallo del Delta +39 06 854511 Cavallo del Ventasso Cavallo Pentro +39 06 85451322 Cavallo Sarcidano Napoletano @ [email protected] Norico-Pinzgauer Persano www www.aia.it Pony di Esperia Sanfratellano Salernitano Tolfetano Asinine: Asino dell’Amiata Asino dell’Asinara Asino di Martina Franca Asino Ragusano Asino Romagnolo Asino Pantesco Asino Sardo AIA-Associazione Italiana 380001 Libro genealogico Murgese Dr. Giancarlo Allevatori cavallo Murgese Carchedi Via Tomassetti, 9 00161 ROMA +39 06 854511 +39 06 85451322 @ [email protected] www www.aia.it ANACRHAI-Associazione 380002 Libro -

Usefulness of the 17-Plex Str Kit for Bosnian Mountain Horse Genotyping
UDC 575. https://doi.org/10.2298/GENSR1902619R Original scientific paper USEFULNESS OF THE 17-PLEX STR KIT FOR BOSNIAN MOUNTAIN HORSE GENOTYPING Dunja RUKAVINA1*, Amir ZAHIROVIĆ2, Ćazim CRNKIĆ3, Mirela MAČKIĆ-ĐUROVIĆ4, Adaleta DURMIĆ-PAŠIĆ5, Belma KALAMUJIĆ STROIL5, Naris POJSKIĆ5 1*University of Sarajevo-Veterinary Faculty, Department of Biology, Sarajevo, B&H 2University of Sarajevo-Veterinary Faculty, Department of Internal Diseases, Sarajevo, B&H 3University of Sarajevo-Veterinary Faculty, Department of Animal Nutrition, Sarajevo, B&H 4University of Sarajevo-Faculty of Medicine, Center for Genetic, Sarajevo, B&H 5University of Sarajevo-Institute for Genetic Engineering and Biotechnology, Sarajevo, B&H Rukavina D., A. Zahirović, Ć. Crnkić, M. Mačkić-Đurović, A. Durmić-Pašić, B. Kalamujić Stroil, N. Pojskić (2019): Usefulness of the 17-plex STR kit for Bosnian mountain horse genotyping.- Genetika, Vol 51, No.2, 619-627. In the present study modern technology of DNA extraction and automatic genotyping was applied in Bosnian and Herzegovinian autochthonous horse breed by using 17-Plex horse genotyping kit. The study was aimed at investigating usefulness of the 17-plex STR Kit for Bosnian mountain horse genotyping and establishing highly useful microsatellite markers system for genetic diversity studies in Bosnian mountain horse breed. Genomic DNA was extracted from whole blood collected from 22 unrelated Bosnian mountain horse specimens. A total of 95 alleles were detected. Average number of detected alleles per locus was 5.588, varying from 3 (HTG7) to 10 (ASB17). Average effective number of alleles was 3.603, fluctuating from 1.789 (HMS7) to 5.728 (HMS2). The observed heterozygosity ranged from 0.136 (HMS3) to 0.909 (ASB2) with a mean of 0.631. -

Selected Readings on the History and Use of Old Livestock Breeds
NATIONAL AGRICULTURAL LIBRARY ARCHIVED FILE Archived files are provided for reference purposes only. This file was current when produced, but is no longer maintained and may now be outdated. Content may not appear in full or in its original format. All links external to the document have been deactivated. For additional information, see http://pubs.nal.usda.gov. Selected Readings on the History and Use of Old Livestock Breeds United States Department of Agriculture Selected Readings on the History and Use of Old Livestock Breeds National Agricultural Library September 1991 Animal Welfare Information Center By: Jean Larson Janice Swanson D'Anna Berry Cynthia Smith Animal Welfare Information Center National Agricultural Library U.S. Department of Agriculture And American Minor Breeds Conservancy P.O. Box 477 Pittboro, NC 27312 Acknowledgement: Jennifer Carter for computer and technical support. Published by: U. S. Department of Agriculture National Agricultural Library Animal Welfare Information Center Beltsville, Maryland 20705 Contact us: http://awic.nal.usda.gov/contact-us Web site: www.nal.usda.gov/awic Published in cooperation with the Virginia-Maryland Regional College of Veterinary Medicine Policies and Links Introduction minorbreeds.htm[1/15/2015 2:16:51 PM] Selected Readings on the History and Use of Old Livestock Breeds For centuries animals have worked with and for people. Cattle, goats, sheep, pigs, poultry and other livestock have been an essential part of agriculture and our history as a nation. With the change of agriculture from a way of life to a successful industry, we are losing our agricultural roots. Although we descend from a nation of farmers, few of us can name more than a handful of livestock breeds that are important to our production of food and fiber.