Novel Insights Into the Assembly and Function of Human Nuclear-Encoded
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cell Development and Peripheral Survival Heme Exporter FLVCR Is
Heme Exporter FLVCR Is Required for T Cell Development and Peripheral Survival Mary Philip, Scott A. Funkhouser, Edison Y. Chiu, Susan R. Phelps, Jeffrey J. Delrow, James Cox, Pamela J. Fink and This information is current as Janis L. Abkowitz of September 28, 2021. J Immunol 2015; 194:1677-1685; Prepublished online 12 January 2015; doi: 10.4049/jimmunol.1402172 http://www.jimmunol.org/content/194/4/1677 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2015/01/09/jimmunol.140217 Material 2.DCSupplemental http://www.jimmunol.org/ References This article cites 44 articles, 11 of which you can access for free at: http://www.jimmunol.org/content/194/4/1677.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 28, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2015 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Heme Exporter FLVCR Is Required for T Cell Development and Peripheral Survival Mary Philip,*,† Scott A. -

Mutation in the COX4I1 Gene Is Associated with Short Stature, Poor Weight Gain and Increased Chromosomal Breaks, Simulating Fanconi Anemia
European Journal of Human Genetics (2017) 25, 1142–1146 & 2017 Macmillan Publishers Limited, part of Springer Nature. All rights reserved 1018-4813/17 www.nature.com/ejhg ARTICLE Mutation in the COX4I1 gene is associated with short stature, poor weight gain and increased chromosomal breaks, simulating Fanconi anemia Bassam Abu-Libdeh*,1,4, Liza Douiev2,3,4, Sarah Amro1, Maher Shahrour1, Asaf Ta-Shma2, Chaya Miller2,3, Orly Elpeleg2 and Ann Saada*,2,3 We describe a novel autosomal recessive form of mitochondrial disease in a child with short stature, poor weight gain, and mild dysmorphic features with highly suspected Fanconi anemia due to a mutation in COX4I1 gene. Whole Exome Sequencing was performed then followed by Sanger confirmation, identified a K101N mutation in COX4I1, segregating with the disease. This nuclear gene encodes the common isoform of cytochrome c oxidase (COX) subunit 4 (COX 4-1), an integral regulatory part of COX (respiratory chain complex IV) the terminal electron acceptor of the mitochondrial respiratory chain. The patient’s fibroblasts disclosed decreased COX activity, impaired ATP production, elevated ROS production, decreased expression of COX4I1 mRNA and undetectable (COX4) protein. COX activity and ATP production were restored by lentiviral transfection with the wild-type gene. Our results demonstrate the first human mutation in the COX4I1 gene linked to diseases and confirm its role in the pathogenesis. Thus COX4I1 mutations should be considered in any patient with features suggestive of this diagnosis. European Journal of Human Genetics (2017) 25, 1142–1146; doi:10.1038/ejhg.2017.112; published online 2 August 2017 INTRODUCTION normal vaginal delivery with birth weight of 2.8 kg after an uneventful Mitochondrial cytochrome c oxidase (COX, complex IV) is the terminal pregnancy. -

Associated 16P11.2 Deletion in Drosophila Melanogaster
ARTICLE DOI: 10.1038/s41467-018-04882-6 OPEN Pervasive genetic interactions modulate neurodevelopmental defects of the autism- associated 16p11.2 deletion in Drosophila melanogaster Janani Iyer1, Mayanglambam Dhruba Singh1, Matthew Jensen1,2, Payal Patel 1, Lucilla Pizzo1, Emily Huber1, Haley Koerselman3, Alexis T. Weiner 1, Paola Lepanto4, Komal Vadodaria1, Alexis Kubina1, Qingyu Wang 1,2, Abigail Talbert1, Sneha Yennawar1, Jose Badano 4, J. Robert Manak3,5, Melissa M. Rolls1, Arjun Krishnan6,7 & 1234567890():,; Santhosh Girirajan 1,2,8 As opposed to syndromic CNVs caused by single genes, extensive phenotypic heterogeneity in variably-expressive CNVs complicates disease gene discovery and functional evaluation. Here, we propose a complex interaction model for pathogenicity of the autism-associated 16p11.2 deletion, where CNV genes interact with each other in conserved pathways to modulate expression of the phenotype. Using multiple quantitative methods in Drosophila RNAi lines, we identify a range of neurodevelopmental phenotypes for knockdown of indi- vidual 16p11.2 homologs in different tissues. We test 565 pairwise knockdowns in the developing eye, and identify 24 interactions between pairs of 16p11.2 homologs and 46 interactions between 16p11.2 homologs and neurodevelopmental genes that suppress or enhance cell proliferation phenotypes compared to one-hit knockdowns. These interac- tions within cell proliferation pathways are also enriched in a human brain-specific network, providing translational relevance in humans. Our study indicates a role for pervasive genetic interactions within CNVs towards cellular and developmental phenotypes. 1 Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA. 2 Bioinformatics and Genomics Program, The Huck Institutes of the Life Sciences, The Pennsylvania State University, University Park, PA 16802, USA. -

Structure of the Intact 14-Subunit Human Cytochrome C Oxidase
www.nature.com/cr www.cell-research.com ARTICLE Structure of the intact 14-subunit human cytochrome c oxidase Shuai Zong 1, Meng Wu1, Jinke Gu1, Tianya Liu1, Runyu Guo1 and Maojun Yang1,2 Respiration is one of the most basic features of living organisms, and the electron transport chain complexes are probably the most complicated protein system in mitochondria. Complex-IV is the terminal enzyme of the electron transport chain, existing either as randomly scattered complexes or as a component of supercomplexes. NDUFA4 was previously assumed as a subunit of complex-I, but recent biochemical data suggested it may be a subunit of complex-IV. However, no structural evidence supporting this notion was available till now. Here we obtained the 3.3 Å resolution structure of complex-IV derived from the human supercomplex I1III2IV1 and assigned the NDUFA4 subunit into complex-IV. Intriguingly, NDUFA4 lies exactly at the dimeric interface observed in previously reported crystal structures of complex-IV homodimer which would preclude complex- IV dimerization. Combining previous structural and biochemical data shown by us and other groups, we propose that the intact complex-IV is a monomer containing 14 subunits. Cell Research (2018) 28:1026–1034; https://doi.org/10.1038/s41422-018-0071-1 INTRODUCTION states under physiological conditions, either being assembled into Mitochondria are critical to many cellular activities. Respiration is supercomplexes or freely scattered on mitochondrial inner the central function of mitochondria, and is exquisitely regulated membrane.36 NDUFA4 was originally considered as a subunit of in multiple ways in response to varying cell conditions.1–3 The Complex-I37 but was proposed to belong to Complex-IV recently.38 assembly of respiratory chain complexes, Complex I–IV (CI, NADH: However, the precise location of NDUFA4 in CIV remains unknown, ubiquinone oxidoreductase; CII, succinate:ubiquinone oxidoreduc- and thus this notion still lacks structural support. -

A High-Resolution Genome-Wide CRISPR/Cas9 Viability Screen
RESEARCH ARTICLE crossm A High-Resolution Genome-Wide CRISPR/Cas9 Viability Screen Reveals Structural Features and Contextual Diversity of the Human Cell-Essential Proteome Downloaded from Thierry Bertomeu,a Jasmin Coulombe-Huntington,a Andrew Chatr-aryamontri,a Karine G. Bourdages,a Etienne Coyaud,b Brian Raught,b Yu Xia,c Mike Tyersa aInstitute for Research in Immunology and Cancer, Department of Medicine, University of Montreal, Montreal, Quebec, Canada bPrincess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada cDepartment of Bioengineering, McGill University, Montreal, Quebec, Canada ABSTRACT To interrogate genes essential for cell growth, proliferation and survival http://mcb.asm.org/ in human cells, we carried out a genome-wide clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 screen in a B-cell lymphoma line using a custom extended-knockout (EKO) library of 278,754 single-guide RNAs (sgRNAs) that tar- geted 19,084 RefSeq genes, 20,852 alternatively spliced exons, and 3,872 hypotheti- cal genes. A new statistical analysis tool called robust analytics and normalization for knockout screens (RANKS) identified 2,280 essential genes, 234 of which were unique. Individual essential genes were validated experimentally and linked to ribo- some biogenesis and stress responses. Essential genes exhibited a bimodal distribu- on December 15, 2018 by guest tion across 10 different cell lines, consistent with a continuous variation in essential- ity as a function of cell type. Genes essential in more lines had more severe fitness defects and encoded the evolutionarily conserved structural cores of protein com- plexes, whereas genes essential in fewer lines formed context-specific modules and encoded subunits at the periphery of essential complexes. -
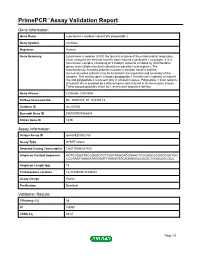
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name cytochrome c oxidase subunit VIa polypeptide 2 Gene Symbol COX6A2 Organism Human Gene Summary Cytochrome c oxidase (COX) the terminal enzyme of the mitochondrial respiratory chain catalyzes the electron transfer from reduced cytochrome c to oxygen. It is a heteromeric complex consisting of 3 catalytic subunits encoded by mitochondrial genes and multiple structural subunits encoded by nuclear genes. The mitochondrially-encoded subunits function in electron transfer and the nuclear-encoded subunits may be involved in the regulation and assembly of the complex. This nuclear gene encodes polypeptide 2 (heart/muscle isoform) of subunit VIa and polypeptide 2 is present only in striated muscles. Polypeptide 1 (liver isoform) of subunit VIa is encoded by a different gene and is found in all non-muscle tissues. These two polypeptides share 66% amino acid sequence identity. Gene Aliases COX6AH, COXVIAH RefSeq Accession No. NC_000016.9, NT_010393.16 UniGene ID Hs.250760 Ensembl Gene ID ENSG00000156885 Entrez Gene ID 1339 Assay Information Unique Assay ID qHsaCED0002180 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000287490 Amplicon Context Sequence GGTGCGGATGCGGAGGTGTTGGTAGGGACGGAACTCGGGGCGCGGGCGGTGG CCCGAGTGGAGATAGGAGTTGAAGGTGCAGAGGGCCACGCTGGGCAGCGCC Amplicon Length (bp) 73 Chromosome Location 16:31439339-31439441 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 98 R2 0.9997 cDNA Cq 28.87 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -

Role of Cytochrome C Oxidase Nuclear-Encoded Subunits in Health and Disease
Physiol. Res. 69: 947-965, 2020 https://doi.org/10.33549/physiolres.934446 REVIEW Role of Cytochrome c Oxidase Nuclear-Encoded Subunits in Health and Disease Kristýna ČUNÁTOVÁ1, David PAJUELO REGUERA1, Josef HOUŠTĚK1, Tomáš MRÁČEK1, Petr PECINA1 1Department of Bioenergetics, Institute of Physiology, Czech Academy of Sciences, Prague, Czech Republic Received February 2, 2020 Accepted September 13, 2020 Epub Ahead of Print November 2, 2020 Summary [email protected] and Tomáš Mráček, Department of Cytochrome c oxidase (COX), the terminal enzyme of Bioenergetics, Institute of Physiology CAS, Vídeňská 1083, 142 mitochondrial electron transport chain, couples electron transport 20 Prague 4, Czech Republic. E-mail: [email protected] to oxygen with generation of proton gradient indispensable for the production of vast majority of ATP molecules in mammalian Cytochrome c oxidase cells. The review summarizes current knowledge of COX structure and function of nuclear-encoded COX subunits, which may Energy demands of mammalian cells are mainly modulate enzyme activity according to various conditions. covered by ATP synthesis carried out by oxidative Moreover, some nuclear-encoded subunits possess tissue-specific phosphorylation apparatus (OXPHOS) located in the and development-specific isoforms, possibly enabling fine-tuning central bioenergetic organelle, mitochondria. OXPHOS is of COX function in individual tissues. The importance of nuclear- composed of five multi-subunit complexes embedded in encoded subunits is emphasized by recently discovered the inner mitochondrial membrane (IMM). Electron pathogenic mutations in patients with severe mitopathies. In transport from reduced substrates of complexes I and II to addition, proteins substoichiometrically associated with COX were cytochrome c oxidase (COX, complex IV, CIV) is found to contribute to COX activity regulation and stabilization of achieved by increasing redox potential of individual the respiratory supercomplexes. -

Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces Cerevisiae
life Review Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae Leticia V. R. Franco 1,2,* , Luca Bremner 1 and Mario H. Barros 2 1 Department of Biological Sciences, Columbia University, New York, NY 10027, USA; [email protected] 2 Department of Microbiology,Institute of Biomedical Sciences, Universidade de Sao Paulo, Sao Paulo 05508-900, Brazil; [email protected] * Correspondence: [email protected] Received: 27 October 2020; Accepted: 19 November 2020; Published: 23 November 2020 Abstract: The ease with which the unicellular yeast Saccharomyces cerevisiae can be manipulated genetically and biochemically has established this organism as a good model for the study of human mitochondrial diseases. The combined use of biochemical and molecular genetic tools has been instrumental in elucidating the functions of numerous yeast nuclear gene products with human homologs that affect a large number of metabolic and biological processes, including those housed in mitochondria. These include structural and catalytic subunits of enzymes and protein factors that impinge on the biogenesis of the respiratory chain. This article will review what is currently known about the genetics and clinical phenotypes of mitochondrial diseases of the respiratory chain and ATP synthase, with special emphasis on the contribution of information gained from pet mutants with mutations in nuclear genes that impair mitochondrial respiration. Our intent is to provide the yeast mitochondrial specialist with basic knowledge of human mitochondrial pathologies and the human specialist with information on how genes that directly and indirectly affect respiration were identified and characterized in yeast. Keywords: mitochondrial diseases; respiratory chain; yeast; Saccharomyces cerevisiae; pet mutants 1. -

COX6A2 Antibody (Center) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap9616c
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 COX6A2 Antibody (Center) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP9616c Specification COX6A2 Antibody (Center) - Product Information Application WB, IHC-P,E Primary Accession Q02221 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Antigen Region 37-66 COX6A2 Antibody (Center) - Additional Information Gene ID 1339 Other Names Cytochrome c oxidase subunit 6A2, mitochondrial, Cytochrome c oxidase Western blot analysis of lysate from human polypeptide VIa-heart, COXVIAH, skeletal muscle tissue lysate, using COX6A2 Cytochrome c oxidase subunit VIA-muscle, Antibody (Center)(Cat. #AP9616c). AP9616c COX VIa-M, COX6A2, COX6A, COX6AH was diluted at 1:1000. A goat anti-rabbit IgG H&L(HRP) at 1:10000 dilution was used as Target/Specificity the secondary antibody. Lysate at 20ug. This COX6A2 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 37-66 amino acids from the Central region of human COX6A2. Dilution WB~~1:1000 IHC-P~~1:25 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Storage Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C Western blot analysis of lysate from human in small aliquots to prevent freeze-thaw heart tissue lysate, using COX6A2 Antibody cycles. (Center)(Cat. #AP9616c). AP9616c was diluted at 1:1000 at each lane. A goat Precautions anti-rabbit IgG H&L(HRP) at 1:5000 dilution Page 1/3 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 COX6A2 Antibody (Center) is for research was used as the secondary antibody. -

Electron Transport Chain Activity Is a Predictor and Target for Venetoclax Sensitivity in Multiple Myeloma
ARTICLE https://doi.org/10.1038/s41467-020-15051-z OPEN Electron transport chain activity is a predictor and target for venetoclax sensitivity in multiple myeloma Richa Bajpai1,7, Aditi Sharma 1,7, Abhinav Achreja2,3, Claudia L. Edgar1, Changyong Wei1, Arusha A. Siddiqa1, Vikas A. Gupta1, Shannon M. Matulis1, Samuel K. McBrayer 4, Anjali Mittal3,5, Manali Rupji 6, Benjamin G. Barwick 1, Sagar Lonial1, Ajay K. Nooka 1, Lawrence H. Boise 1, Deepak Nagrath2,3,5 & ✉ Mala Shanmugam 1 1234567890():,; The BCL-2 antagonist venetoclax is highly effective in multiple myeloma (MM) patients exhibiting the 11;14 translocation, the mechanistic basis of which is unknown. In evaluating cellular energetics and metabolism of t(11;14) and non-t(11;14) MM, we determine that venetoclax-sensitive myeloma has reduced mitochondrial respiration. Consistent with this, low electron transport chain (ETC) Complex I and Complex II activities correlate with venetoclax sensitivity. Inhibition of Complex I, using IACS-010759, an orally bioavailable Complex I inhibitor in clinical trials, as well as succinate ubiquinone reductase (SQR) activity of Complex II, using thenoyltrifluoroacetone (TTFA) or introduction of SDHC R72C mutant, independently sensitize resistant MM to venetoclax. We demonstrate that ETC inhibition increases BCL-2 dependence and the ‘primed’ state via the ATF4-BIM/NOXA axis. Further, SQR activity correlates with venetoclax sensitivity in patient samples irrespective of t(11;14) status. Use of SQR activity in a functional-biomarker informed manner may better select for MM patients responsive to venetoclax therapy. 1 Department of Hematology and Medical Oncology, Winship Cancer Institute, School of Medicine, Emory University, Atlanta, GA, USA. -

The Overexpression of Cytochrome C Oxidase Subunit 6C Activated by Kras Mutation Is Related to Energy Metabolism in Pancreatic Cancer
300 Original Article The overexpression of cytochrome c oxidase subunit 6C activated by Kras mutation is related to energy metabolism in pancreatic cancer Jigang Yang1, Jun Liu1, Shuxin Zhang1, Yuanyuan Yang1, Jianhua Gong2 1Department of Nuclear Medicine, Beijing Friendship Hospital, affiliated to Capital Medical University, Beijing 100050, China; 2Department of Oncology, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, China. Contributions: (I) Conception and design: J Yang, J Gong; (II) Administrative support: J Gong; (III) Provision of study materials: J Liu; (IV) Collection and assembly of data: S Zhang, Y Yang; (V) Data analysis and interpretation: S Zhang, Y Yang; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Jianhua Gong. Oncology department, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences, 1# Tiantan Xili, Beijing 100050, China. Email: [email protected]. Background: Kras mutation is frequently detected in pancreatic cancers and leads to altered energy metabolite. Here we investigated molecule markers related with Kras mutation, which could be used as developing new target for Kras mutant driven cancer. Methods: A knockin BxPC-3/KrasG12D cell line was constructed by CRISPR/Cas9 system. Proliferation and metabolite characterization in BxPC-3/KrasG12D was compared with wild type BxPC-3 by using colony formation assay and mitochondrial dyes. The differential genes were screened using mitochondrial metabolite-related genes PCR array. The expression of COX6C was confirmed by real time polymerase chain reaction (RT-PCR) and western blot. COX6C expression in 30 pairs of tissue microarray of pancreatic carcinoma and matched adjacent tissues was analyzed by immunohistochemistry.