Cell Development and Peripheral Survival Heme Exporter FLVCR Is
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Associated 16P11.2 Deletion in Drosophila Melanogaster
ARTICLE DOI: 10.1038/s41467-018-04882-6 OPEN Pervasive genetic interactions modulate neurodevelopmental defects of the autism- associated 16p11.2 deletion in Drosophila melanogaster Janani Iyer1, Mayanglambam Dhruba Singh1, Matthew Jensen1,2, Payal Patel 1, Lucilla Pizzo1, Emily Huber1, Haley Koerselman3, Alexis T. Weiner 1, Paola Lepanto4, Komal Vadodaria1, Alexis Kubina1, Qingyu Wang 1,2, Abigail Talbert1, Sneha Yennawar1, Jose Badano 4, J. Robert Manak3,5, Melissa M. Rolls1, Arjun Krishnan6,7 & 1234567890():,; Santhosh Girirajan 1,2,8 As opposed to syndromic CNVs caused by single genes, extensive phenotypic heterogeneity in variably-expressive CNVs complicates disease gene discovery and functional evaluation. Here, we propose a complex interaction model for pathogenicity of the autism-associated 16p11.2 deletion, where CNV genes interact with each other in conserved pathways to modulate expression of the phenotype. Using multiple quantitative methods in Drosophila RNAi lines, we identify a range of neurodevelopmental phenotypes for knockdown of indi- vidual 16p11.2 homologs in different tissues. We test 565 pairwise knockdowns in the developing eye, and identify 24 interactions between pairs of 16p11.2 homologs and 46 interactions between 16p11.2 homologs and neurodevelopmental genes that suppress or enhance cell proliferation phenotypes compared to one-hit knockdowns. These interac- tions within cell proliferation pathways are also enriched in a human brain-specific network, providing translational relevance in humans. Our study indicates a role for pervasive genetic interactions within CNVs towards cellular and developmental phenotypes. 1 Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA. 2 Bioinformatics and Genomics Program, The Huck Institutes of the Life Sciences, The Pennsylvania State University, University Park, PA 16802, USA. -

Structure of the Intact 14-Subunit Human Cytochrome C Oxidase
www.nature.com/cr www.cell-research.com ARTICLE Structure of the intact 14-subunit human cytochrome c oxidase Shuai Zong 1, Meng Wu1, Jinke Gu1, Tianya Liu1, Runyu Guo1 and Maojun Yang1,2 Respiration is one of the most basic features of living organisms, and the electron transport chain complexes are probably the most complicated protein system in mitochondria. Complex-IV is the terminal enzyme of the electron transport chain, existing either as randomly scattered complexes or as a component of supercomplexes. NDUFA4 was previously assumed as a subunit of complex-I, but recent biochemical data suggested it may be a subunit of complex-IV. However, no structural evidence supporting this notion was available till now. Here we obtained the 3.3 Å resolution structure of complex-IV derived from the human supercomplex I1III2IV1 and assigned the NDUFA4 subunit into complex-IV. Intriguingly, NDUFA4 lies exactly at the dimeric interface observed in previously reported crystal structures of complex-IV homodimer which would preclude complex- IV dimerization. Combining previous structural and biochemical data shown by us and other groups, we propose that the intact complex-IV is a monomer containing 14 subunits. Cell Research (2018) 28:1026–1034; https://doi.org/10.1038/s41422-018-0071-1 INTRODUCTION states under physiological conditions, either being assembled into Mitochondria are critical to many cellular activities. Respiration is supercomplexes or freely scattered on mitochondrial inner the central function of mitochondria, and is exquisitely regulated membrane.36 NDUFA4 was originally considered as a subunit of in multiple ways in response to varying cell conditions.1–3 The Complex-I37 but was proposed to belong to Complex-IV recently.38 assembly of respiratory chain complexes, Complex I–IV (CI, NADH: However, the precise location of NDUFA4 in CIV remains unknown, ubiquinone oxidoreductase; CII, succinate:ubiquinone oxidoreduc- and thus this notion still lacks structural support. -
Primepcr™Assay Validation Report
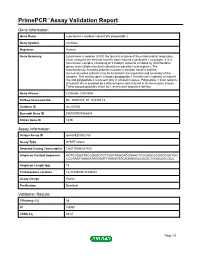
PrimePCR™Assay Validation Report Gene Information Gene Name cytochrome c oxidase subunit VIa polypeptide 2 Gene Symbol COX6A2 Organism Human Gene Summary Cytochrome c oxidase (COX) the terminal enzyme of the mitochondrial respiratory chain catalyzes the electron transfer from reduced cytochrome c to oxygen. It is a heteromeric complex consisting of 3 catalytic subunits encoded by mitochondrial genes and multiple structural subunits encoded by nuclear genes. The mitochondrially-encoded subunits function in electron transfer and the nuclear-encoded subunits may be involved in the regulation and assembly of the complex. This nuclear gene encodes polypeptide 2 (heart/muscle isoform) of subunit VIa and polypeptide 2 is present only in striated muscles. Polypeptide 1 (liver isoform) of subunit VIa is encoded by a different gene and is found in all non-muscle tissues. These two polypeptides share 66% amino acid sequence identity. Gene Aliases COX6AH, COXVIAH RefSeq Accession No. NC_000016.9, NT_010393.16 UniGene ID Hs.250760 Ensembl Gene ID ENSG00000156885 Entrez Gene ID 1339 Assay Information Unique Assay ID qHsaCED0002180 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000287490 Amplicon Context Sequence GGTGCGGATGCGGAGGTGTTGGTAGGGACGGAACTCGGGGCGCGGGCGGTGG CCCGAGTGGAGATAGGAGTTGAAGGTGCAGAGGGCCACGCTGGGCAGCGCC Amplicon Length (bp) 73 Chromosome Location 16:31439339-31439441 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 98 R2 0.9997 cDNA Cq 28.87 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -

INVESTIGATION INTO SUBGROUP C Felv INTERACTION with ITS HOST RECEPTOR FLVCR1 and the ROLE of FLVCR1 in DIAMOND BLACKFAN ANEMIA
INVESTIGATION INTO SUBGROUP C FeLV INTERACTION WITH ITS HOST RECEPTOR FLVCR1 AND THE ROLE OF FLVCR1 IN DIAMOND BLACKFAN ANEMIA By Michelle Antoinette Rey A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Graduate Department of Molecular Genetics University of Toronto © Copyright by Michelle Antoinette Rey 2009 Investigation in subgroup C FeLV interaction with its host receptor FLVCR1 And the role of FLVCR1 in Diamond Blackfan Anemia For the degree of Doctor of Philosophy, 2009 By Michelle Antoinette Rey Graduate Department of Molecular Genetics University of Toronto ABSTRACT Retroviral infection requires an initial interaction between the host cell and the virion. This interaction is predominantly mediated by an envelope (env) protein exposed on the external face of the virion. For gammaretroviruses, such as feline leukemia virus (FeLV), the receptor-binding domain (RBD) is located in the N terminus of env. The RBD forms a distinct domain that is sufficient for binding to the host receptor, but is inefficient in the absence of the corresponding C terminal env, Cdomain, sequence in viral infection studies. I developed a series of hybrid constructs between subgroup C, A and T FeLVs that use distinct receptors for infection to determine the role of Cdom in FeLV binding and infection. Using this approach, I have shown that the C domain (Cdom) of FeLV-C env forms a second receptor-binding domain, distinct from its RBD, which is critical for efficient binding and infection of FeLV-C to host cells expressing FLVCR1. I propose that this mechanism of interaction is conserved for all gammaretroviruses. -

1589467268 490 16.Pdf
Veterinary Microbiology 229 (2019) 147–152 Contents lists available at ScienceDirect Veterinary Microbiology journal homepage: www.elsevier.com/locate/vetmic Expression of the feline leukemia virus subgroup C receptors in normal and neoplastic urothelium of the urinary bladder of cattle associated with bovine T papillomavirus infection Valeria Russoa, Franco Ropertob, Marian Taulescuc, Francesca De Falcoa, Chiara Urraroa, ⁎ Federica Corradod, John S. Mundaye, Cornel Catoic, Sante Ropertoa, a Dipartimento di Medicina Veterinaria e Produzioni Animali, Università di Napoli Federico II, Napoli, Italy b Dipartimento di Biologia, Università di Napoli Federico II, Napoli, Italy c University of Agricultural Sciences and Veterinary Medicine, Faculty of Veterinary Medicine, Pathology Department, Cluj-Napoca, Romania d Istituto Zooprofilattico Sperimentale del Mezzogiorno, Portici, Napoli, Italy e Pathobiology, School of Veterinary Sciences, Massey University, Palmerston North, New Zealand ARTICLE INFO ABSTRACT Keywords: The feline leukemia virus subgroup C receptors (FLVCRs) were originally cloned as virus receptors, but are now Bovine papillomavirus type 2 (BPV2) believed to function also as heme transporters and are expressed in a broad range of tissues in a wide range of BPV13 mammalian species. Expression of FLVCR1 and FLVCR2 was investigated in 19 bovine papillomavirus-associated Deltapapillomavirus urinary bladder cancers and in 15 non-neoplastic samples of bladder from cattle. E5 oncoprotein of bovine Feline leukemia virus subgroup C receptror1 Deltapapillomaviruses (δPVs) was detected in 17 of the 19 bladder cancers. Flvcr1 and Flvcr2 were amplified (FLVCR1) and sequenced both in neoplastic and non-neoplastic samples showing a 100% identity with bovine Flvcr1 and FLVCR2 Urothelial cancer Flvcr2 mRNA sequences present in GenBank database (accession numbers: NM_001206019.1 and NM_001192143.1, respectively). -

COX6A2 Antibody (Center) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap9616c
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 COX6A2 Antibody (Center) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP9616c Specification COX6A2 Antibody (Center) - Product Information Application WB, IHC-P,E Primary Accession Q02221 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Antigen Region 37-66 COX6A2 Antibody (Center) - Additional Information Gene ID 1339 Other Names Cytochrome c oxidase subunit 6A2, mitochondrial, Cytochrome c oxidase Western blot analysis of lysate from human polypeptide VIa-heart, COXVIAH, skeletal muscle tissue lysate, using COX6A2 Cytochrome c oxidase subunit VIA-muscle, Antibody (Center)(Cat. #AP9616c). AP9616c COX VIa-M, COX6A2, COX6A, COX6AH was diluted at 1:1000. A goat anti-rabbit IgG H&L(HRP) at 1:10000 dilution was used as Target/Specificity the secondary antibody. Lysate at 20ug. This COX6A2 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 37-66 amino acids from the Central region of human COX6A2. Dilution WB~~1:1000 IHC-P~~1:25 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Storage Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C Western blot analysis of lysate from human in small aliquots to prevent freeze-thaw heart tissue lysate, using COX6A2 Antibody cycles. (Center)(Cat. #AP9616c). AP9616c was diluted at 1:1000 at each lane. A goat Precautions anti-rabbit IgG H&L(HRP) at 1:5000 dilution Page 1/3 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 COX6A2 Antibody (Center) is for research was used as the secondary antibody. -

The Overexpression of Cytochrome C Oxidase Subunit 6C Activated by Kras Mutation Is Related to Energy Metabolism in Pancreatic Cancer
300 Original Article The overexpression of cytochrome c oxidase subunit 6C activated by Kras mutation is related to energy metabolism in pancreatic cancer Jigang Yang1, Jun Liu1, Shuxin Zhang1, Yuanyuan Yang1, Jianhua Gong2 1Department of Nuclear Medicine, Beijing Friendship Hospital, affiliated to Capital Medical University, Beijing 100050, China; 2Department of Oncology, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, China. Contributions: (I) Conception and design: J Yang, J Gong; (II) Administrative support: J Gong; (III) Provision of study materials: J Liu; (IV) Collection and assembly of data: S Zhang, Y Yang; (V) Data analysis and interpretation: S Zhang, Y Yang; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Jianhua Gong. Oncology department, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences, 1# Tiantan Xili, Beijing 100050, China. Email: [email protected]. Background: Kras mutation is frequently detected in pancreatic cancers and leads to altered energy metabolite. Here we investigated molecule markers related with Kras mutation, which could be used as developing new target for Kras mutant driven cancer. Methods: A knockin BxPC-3/KrasG12D cell line was constructed by CRISPR/Cas9 system. Proliferation and metabolite characterization in BxPC-3/KrasG12D was compared with wild type BxPC-3 by using colony formation assay and mitochondrial dyes. The differential genes were screened using mitochondrial metabolite-related genes PCR array. The expression of COX6C was confirmed by real time polymerase chain reaction (RT-PCR) and western blot. COX6C expression in 30 pairs of tissue microarray of pancreatic carcinoma and matched adjacent tissues was analyzed by immunohistochemistry. -

Product Sheet CP10253
COX4I1 Antibody Applications: WB, IP, ICC, FACS Detected MW: 19 kDa Cat. No. CP10253 Species & Reactivity: Human, Mouse, Rat Isotype: Mouse IgG1 BACKGROUND Cytochrome c oxidase (EC 1.9.3.1) (COX) belongs from invertebrates to mammals, in most cases to the superfamily of terminal oxidases present in mimic the pathological features of the human all aerobic organisms. In eukaryotic cells, diseases.3 cytochrome c oxidase is embedded as a dimer in the mitochondrial inner membrane. Electron References transfer by the enzyme from cytochrome c to 1. Capaldi, R.A.: Annu. Rev. Biochem. 59: 569-596, 1990 molecular oxygen is coupled to proton pumping 2. Pecina, P. et al: Physiol. Res. 53(Suppl. 1):S213-S223, across the membrane, contributing to the 2004 mitochondrial membrane potential, resulting in a 3. Diaz, F.: Biochim Biophys Acta. 1802:100-10, 2010 proton and charge gradient that is then employed by the F0F1-ATPase to synthesize ATP. The redox TECHNICAL INFORMATION centers involved in electron transfer are two copper centers (CuA and CuB) and two heme A Source: moieties (a and a3). The eukaryotic enzyme COX4I1 Antibody is a mouse monoclonal antibody contains three mitochondrial DNA (mtDNA)- raised against purified recombinant human COX4I1 encoded subunits: MTCO1, MTCO2, and MTCO3. fragments expressed in E. coli. These subunits are homologous to polypeptides found in bacterial cytochrome c oxidases. They Specificity and Sensitivity: form both the catalytic and structural core of the This antibody detects endogenous COX4I1 proteins enzyme. The hydrophobic interior of enzyme without cross-reactivity with other related subunit 1 (MTCO1) coordinates the heme a group proteins. -

Human Primitive Brain Displays Negative Mitochondrial-Nuclear Expression Correlation of Respiratory Genes
Downloaded from genome.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press Research Human primitive brain displays negative mitochondrial-nuclear expression correlation of respiratory genes Gilad Barshad, Amit Blumberg, Tal Cohen, and Dan Mishmar Department of Life Sciences, Ben-Gurion University of the Negev, Beer Sheva 8410501, Israel Oxidative phosphorylation (OXPHOS), a fundamental energy source in all human tissues, requires interactions between mitochondrial (mtDNA)- and nuclear (nDNA)-encoded protein subunits. Although such interactions are fundamental to OXPHOS, bi-genomic coregulation is poorly understood. To address this question, we analyzed ∼8500 RNA-seq exper- iments from 48 human body sites. Despite well-known variation in mitochondrial activity, quantity, and morphology, we found overall positive mtDNA-nDNA OXPHOS genes’ co-expression across human tissues. Nevertheless, negative mtDNA- nDNA gene expression correlation was identified in the hypothalamus, basal ganglia, and amygdala (subcortical brain re- gions, collectively termed the “primitive” brain). Single-cell RNA-seq analysis of mouse and human brains revealed that this phenomenon is evolutionarily conserved, and both are influenced by brain cell types (involving excitatory/inhibitory neu- rons and nonneuronal cells) and by their spatial brain location. As the “primitive” brain is highly oxidative, we hypothesized that such negative mtDNA-nDNA co-expression likely controls for the high mtDNA transcript levels, which enforce tight OXPHOS regulation, rather than rewiring toward glycolysis. Accordingly, we found “primitive” brain-specific up-regula- tion of lactate dehydrogenase B (LDHB), which associates with high OXPHOS activity, at the expense of LDHA, which pro- motes glycolysis. Analyses of co-expression, DNase-seq, and ChIP-seq experiments revealed candidate RNA-binding proteins and CEBPB as the best regulatory candidates to explain these phenomena. -

Oxidative-Phosphorylation Genes from the Mitochondrial and Nuclear
bioRxiv preprint doi: https://doi.org/10.1101/136457; this version posted May 11, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Oxidative-phosphorylation genes from the mitochondrial and nuclear genomes co-express in all human tissues except for the ancient brain Gilad Barshad1, Amit Blumberg1 and Dan Mishmar1* Department of Life Sciences, Ben-Gurion University of the Negev, Beer Sheva 8410501, Israel *Corresponding author Address: Dan Mishmar, PhD Department of Life Sciences Ben-Gurion University of the Negev Beer Sheva 8410501, Israel Tel: +972-86461355 FAX: 972-86461356 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/136457; this version posted May 11, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract In humans, oxidative phosphorylation (OXPHOS), the cellular energy producer, harbors ~90 nuclear DNA (nDNA)- and mitochondrial DNA (mtDNA)-encoded subunits. Although nDNA- and mtDNA-encoded OXPHOS proteins physically interact, their transcriptional regulation profoundly diverges, thus questioning their co-regulation. To address mtDNA-nDNA gene co- expression, we analyzed ~8,500 RNA-seq Gene-Tissue-Expression (GTEx) experiments encompassing 48 human tissues. We found overall positive cross-tissue mtDNA-nDNA OXPHOS gene co-expression.