Advances in Cytogenetics of Brazilian Rodents: Cytotaxonomy, Chromosome Evolution and New Karyotypic Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sinopsis De Los Mamíferos Silvestres Del Estado De Guanajuato, México, Y Comentarios Sobre Su Conservación Óscar Sánchez 369
La Portada Fotografía tomada en la comida de celebración posterior a la firma del Acta Constitutiva de la Asociación Mexicana de Mastozoología Asociación Civil (AMMAC). La primera mesa directiva de la AMMAC estuvo constituida por Juan Pablo Gallo como presidente, Daniel Navarro como vicepresidente, Rodrigo Medellín como secretario ejecutivo y María Canela como tesorera. La foto fue tomada en casa de Juan Pablo Gallo, que junto con la de Rodrigo Medellín fueron las que hospedaron la mayor cantidad de reuniones preparatorias para la Asociación. De pie empezando por la izquierda: Daniel Navarro, Rosario Manzanos, María Canela, Silvia Manzanilla, Esther Romo, Livia León, Alondra Castro. Sentados desde la izquierda: Federico Romero, Héctor Arita, Rodrigo Medellín, Hiram Barrios, Víctor Sánchez Cordero, Juan Pablo Gallo y Álvaro Miranda (foto tomada por Agustín Gallo Reynoso). Nuestro logo “Ozomatli” El nombre de “Ozomatli” proviene del náhuatl se refiere al símbolo astrológico del mono en el calendario azteca, así como al dios de la danza y del fuego. Se relaciona con la alegría, la danza, el canto, las habilidades. Al signo decimoprimero en la cosmogonía mexica. “Ozomatli” es una representación pictórica de los mono arañas (Ateles geoffroyi). La especie de primate de más amplia distribución en México. “Es habitante de los bosques, sobre todo de los que están por donde sale el sol en Anáhuac. Tiene el dorso pequeño, es barrigudo y su cola, que a veces se enrosca, es larga. Sus manos y sus pies parecen de hombre; también sus uñas. Los Ozomatin gritan y silban y hacen visajes a la gente. Arrojan piedras y palos. -

Riqueza De Espécies E Relevância Para a Conservação
O Brasil é reconhecidamente um dos países de megadiversidade de mamíferos do mundo, abrigando cerca de 12% de todas as espécies desse grupo existentes no nosso planeta, distribuídas em 12 Ordens e 50 Famílias. Dentre as espécies que ocorrem no País, 210 (30% do total) são exclusivas do território brasileiro. Esses números não só indicam a importância do País para a conservação mundial desses animais como também trazem para a mastozoologia brasileira a responsabilidade de produzir e disseminar conhecimento científico de qualidade sobre um grupo carismático, bastante ameaçado pela ação antrópica e importante componente dos ecossistemas naturais. O próprio aumento no número de espécies reconhecidas para o Brasil nos últimos 15 anos já é um indicativo da resposta que vem sendo dada pelos pesquisadores do País a esse desafio de gerar conhecimento científico de qualidade sobre os mamíferos. Na publicação pioneira de Fonseca e colaboradores (Lista Anotada dos Mamíferos do Brasil, 1996), houve a indicação de 524 espécies brasileiras de mamíferos. Na compilação mais recente, de 2012, esse número passou para 701, o que representa um aumento de quase 34% em 16 anos (Paglia et al., Lista Anotada dos Mamíferos do Brasil , 2a ed., 2012). Visando contribuir para essa produção de conhecimento científico de qualidade sobre mamíferos, há alguns anos atrás nós organizamos uma publicação que reunia estudos científicos inéditos sobre vários aspectos da biologia do grupo, intitulada Mamíferos do Brasil: Genética, Sistemática, Ecologia e Conservação. Esse livro, publicado em 2006, contou com a participação de vários mastozoólogos brasileiros de destaque. A nossa intenção, com o mesmo, era contribuir para a produção e divulgação da informação científica para um público mais amplo, incluindo alunos de graduação e não-acadêmicos interessados em mastozoologia, além é claro dos pesquisadores especialistas na área. -

The Pattern of Color Change in Small Mammal
Sandoval Salinas et al. BMC Res Notes (2018) 11:424 https://doi.org/10.1186/s13104-018-3544-x BMC Research Notes RESEARCH NOTE Open Access The pattern of color change in small mammal museum specimens: is it independent of storage histories given museum‑specifc conditions? María Leonor Sandoval Salinas1,2*, José D. Sandoval1,3, Elisa M. Colombo1,3 and Rubén M. Barquez2 Abstract Objective: Determination of color and evaluating its variation form the basis for a broad range of research questions. For studies on taxonomy, systematics, etc., resorting to mammal specimens in museum collections has a number of advantages over using feld specimens. However, if museum specimens are to be for studying color, they should accurately represent the color of live animals, or we should understand how they difer. Basically, this study addresses this question: How does coat color vary when dealing with specimens of Akodon budini (Budin’s grass mouse, Thomas 1918), stored in one museum collection for diferent periods of time? Results: We measured color values through a spectroradiometer and a difuse illumination cabin and used the refectance values in the form of CIELab tri-stimulus values, considering CIE standard illuminant A. We observed that there is a relationship between specimen storage antiquity and pelage color and it seems that it is general for at least a number of small mammals and this could indicate a universal phenomenon across several mammal species and across several storage conditions. Our results, as others, emphasize the importance of considering storage time, among other circumstances, in research studies using mammal skins and where color is of importance. -
Supporting Files
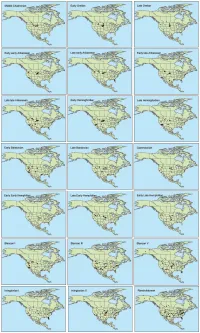
Table S1. Summary of Special Emissions Report Scenarios (SERs) to which we fit climate models for extant mammalian species. Mean Annual Temperature Standard Scenario year (˚C) Deviation Standard Error Present 4.447 15.850 0.057 B1_low 2050s 5.941 15.540 0.056 B1 2050s 6.926 15.420 0.056 A1b 2050s 7.602 15.336 0.056 A2 2050s 8.674 15.163 0.055 A1b 2080s 7.390 15.444 0.056 A2 2080s 9.196 15.198 0.055 A2_top 2080s 11.225 14.721 0.053 Table S2. List of mammalian taxa included and excluded from the species distribution models. -

The Neotropical Region Sensu the Areas of Endemism of Terrestrial Mammals
Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Supplementary material The Neotropical region sensu the areas of endemism of terrestrial mammals Elkin Alexi Noguera-UrbanoA,B,C,D and Tania EscalanteB APosgrado en Ciencias Biológicas, Unidad de Posgrado, Edificio A primer piso, Circuito de Posgrados, Ciudad Universitaria, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. BGrupo de Investigación en Biogeografía de la Conservación, Departamento de Biología Evolutiva, Facultad de Ciencias, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. CGrupo de Investigación de Ecología Evolutiva, Departamento de Biología, Universidad de Nariño, Ciudadela Universitaria Torobajo, 1175-1176 Nariño, Colombia. DCorresponding author. Email: [email protected] Page 1 of 18 Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Table S1. List of taxa processed Number Taxon Number Taxon 1 Abrawayaomys ruschii 55 Akodon montensis 2 Abrocoma 56 Akodon mystax 3 Abrocoma bennettii 57 Akodon neocenus 4 Abrocoma boliviensis 58 Akodon oenos 5 Abrocoma budini 59 Akodon orophilus 6 Abrocoma cinerea 60 Akodon paranaensis 7 Abrocoma famatina 61 Akodon pervalens 8 Abrocoma shistacea 62 Akodon philipmyersi 9 Abrocoma uspallata 63 Akodon reigi 10 Abrocoma vaccarum 64 Akodon sanctipaulensis 11 Abrocomidae 65 Akodon serrensis 12 Abrothrix 66 Akodon siberiae 13 Abrothrix andinus 67 Akodon simulator 14 Abrothrix hershkovitzi 68 Akodon spegazzinii 15 Abrothrix illuteus -

Chromatic Anomalies in Akodontini (Cricetidae: Sigmodontinae) F
Brazilian Journal of Biology https://doi.org/10.1590/1519-6984.214680 ISSN 1519-6984 (Print) Notes and Comments ISSN 1678-4375 (Online) Chromatic anomalies in Akodontini (Cricetidae: Sigmodontinae) F. A. Silvaa,b* , G. Lessab , F. Bertuolc , T. R. O. Freitasd,e and F. M. Quintelaf aPrograma de Pós-graduação em Ecologia e Conservação, Instituto de Biociências, Universidade Federal de Mato Grosso do Sul – UFMS, Cidade Universitária, CEP 79070-900, Campo Grande, MS, Brasil bLaboratório de Mastozoologia, Programa de Pós-graduação em Biologia Animal, Museu de Zoologia João Moojen, Universidade Federal de Viçosa – UFV, Vila Gianetti, Casa 32, Campus Universitário, CEP 36571-000, Viçosa, MG, Brasil cLaboratório de Evolução e Genética Animal, Programa de Pós-graduação em Genética, Conservação e Biologia Evolutiva, Universidade Federal do Amazonas – UFAM, Av. General Rodrigo Otávio Jordão Ramos, 3000, CEP 69077-000, Manaus, AM, Brasil dPrograma de Pós-graduação em Genética e Biologia Molecular, Universidade Federal do Rio Grande do Sul – UFRGS, Av. Bento Gonçalves, 1500, CEP 90040-060, Porto Alegre, RS, Brasil ePrograma de Pós-graduação em Biologia Animal, Departamento de Genética, Universidade Federal do Rio Grande do Sul – UFRGS, Av. Bento Gonçalves, 1500, CEP 90040-060, Porto Alegre, RS, Brasil fLaboratório de Vertebrados, Programa de Pós-graduação em Biologia de Ambientes Aquáticos Continentais, Universidade Federal do Rio Grande – FURG, Av. Itália, CEP 96203-900, Rio Grande, RS, Brasil *e-mail: [email protected] Received: September 27, 2018 – Accepted: February 15, 2019 – Distributed: May 31, 2020 (With 1 figure) The patterns and variability of colors in extant and Álvarez-León, 2014), birds (e. g. -

Cricetidae, Sigmodontinae): Searching for Ancestral Phylogenetic Traits
RESEARCH ARTICLE Extensive Chromosomal Reorganization in the Evolution of New World Muroid Rodents (Cricetidae, Sigmodontinae): Searching for Ancestral Phylogenetic Traits Adenilson Leão Pereira1, Stella Miranda Malcher1, Cleusa Yoshiko Nagamachi1,2, Patricia Caroline Mary O’Brien3, Malcolm Andrew Ferguson-Smith3, Ana Cristina Mendes- Oliveira4, Julio Cesar Pieczarka1,2* 1 Laboratório de Citogenética, Centro de Estudos Avançados da Biodiversidade, ICB, Universidade Federal do Pará, Belém, Pará, Brasil, 2 CNPq Researcher, Brasília, Brasil, 3 Cambridge Resource Center for Comparative Genomics, Department of Veterinary Medicine, University of Cambridge, Cambridge, United Kingdom, 4 Laboratório de Zoologia e Ecologia de Vertebrados, ICB, Universidade Federal do Pará, Belém, Pará, Brasil * [email protected] OPEN ACCESS Citation: Pereira AL, Malcher SM, Nagamachi CY, O’Brien PCM, Ferguson-Smith MA, Mendes-Oliveira Abstract AC, et al. (2016) Extensive Chromosomal Reorganization in the Evolution of New World Muroid Sigmodontinae rodents show great diversity and complexity in morphology and ecology. Rodents (Cricetidae, Sigmodontinae): Searching for This diversity is accompanied by extensive chromosome variation challenging attempts to Ancestral Phylogenetic Traits. PLoS ONE 11(1): reconstruct their ancestral genome. The species Hylaeamys megacephalus–HME (Oryzo- e0146179. doi:10.1371/journal.pone.0146179 myini, 2n = 54), Necromys lasiurus—NLA (Akodontini, 2n = 34) and Akodon sp.–ASP (Ako- Editor: Riccardo Castiglia, Universita degli Studi di dontini, 2n = 10) have extreme diploid numbers that make it difficult to understand the Roma La Sapienza, ITALY rearrangements that are responsible for such differences. In this study we analyzed these Received: June 5, 2015 changes using whole chromosome probes of HME in cross-species painting of NLA and Accepted: December 13, 2015 ASP to construct chromosome homology maps that reveal the rearrangements between Published: January 22, 2016 species. -

With Focus on the Genus Handleyomys and Related Taxa
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2015-04-01 Evolution and Biogeography of Mesoamerican Small Mammals: With Focus on the Genus Handleyomys and Related Taxa Ana Villalba Almendra Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Biology Commons BYU ScholarsArchive Citation Villalba Almendra, Ana, "Evolution and Biogeography of Mesoamerican Small Mammals: With Focus on the Genus Handleyomys and Related Taxa" (2015). Theses and Dissertations. 5812. https://scholarsarchive.byu.edu/etd/5812 This Dissertation is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Evolution and Biogeography of Mesoamerican Small Mammals: Focus on the Genus Handleyomys and Related Taxa Ana Laura Villalba Almendra A dissertation submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Doctor of Philosophy Duke S. Rogers, Chair Byron J. Adams Jerald B. Johnson Leigh A. Johnson Eric A. Rickart Department of Biology Brigham Young University March 2015 Copyright © 2015 Ana Laura Villalba Almendra All Rights Reserved ABSTRACT Evolution and Biogeography of Mesoamerican Small Mammals: Focus on the Genus Handleyomys and Related Taxa Ana Laura Villalba Almendra Department of Biology, BYU Doctor of Philosophy Mesoamerica is considered a biodiversity hot spot with levels of endemism and species diversity likely underestimated. For mammals, the patterns of diversification of Mesoamerican taxa still are controversial. Reasons for this include the region’s complex geologic history, and the relatively recent timing of such geological events. -

Novltatesamerican MUSEUM PUBLISHED by the AMERICAN MUSEUM of NATURAL HISTORY CENTRAL PARK WEST at 79TH STREET, NEW YORK, N.Y
NovltatesAMERICAN MUSEUM PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, N.Y. 10024 Number 3085, 39 pp., 17 figures, 6 tables December 27, 1993 A New Genus for Hesperomys molitor Winge and Holochilus magnus Hershkovitz (Mammalia, Muridae) with an Analysis of Its Phylogenetic Relationships ROBERT S. VOSS1 AND MICHAEL D. CARLETON2 CONTENTS Abstract ............................................. 2 Resumen ............................................. 2 Resumo ............................................. 3 Introduction ............................................. 3 Acknowledgments ............... .............................. 4 Materials and Methods ..................... ........................ 4 Lundomys, new genus ............... .............................. 5 Lundomys molitor (Winge, 1887) ............................................. 5 Comparisons With Holochilus .............................................. 11 External Morphology ................... ........................... 13 Cranium and Mandible ..................... ........................ 15 Dentition ............................................. 19 Viscera ............................................. 20 Phylogenetic Relationships ....................... ...................... 21 Character Definitions ................... .......................... 23 Results .............................................. 27 Phylogenetic Diagnosis and Contents of Oryzomyini ........... .................. 31 Natural History and Zoogeography -

Parasitizing Akodon Montensis (Rodentia: Cricetidae) in the Southern Region of Brazil Revista Brasileira De Parasitologia Veterinária, Vol
Revista Brasileira de Parasitologia Veterinária ISSN: 0103-846X [email protected] Colégio Brasileiro de Parasitologia Veterinária Brasil Trevisan Gressler, Lucas; da Silva Krawczak, Felipe; Knoff, Marcelo; Gonzalez Monteiro, Silvia; Bahia Labruna, Marcelo; de Campos Binder, Lina; Sobotyk de Oliveira, Caroline; Notarnicola, Juliana Litomosoides silvai (Nematoda: Onchocercidae) parasitizing Akodon montensis (Rodentia: Cricetidae) in the southern region of Brazil Revista Brasileira de Parasitologia Veterinária, vol. 26, núm. 4, octubre, 2017, pp. 433- 438 Colégio Brasileiro de Parasitologia Veterinária Jaboticabal, Brasil Available in: http://www.redalyc.org/articulo.oa?id=397853594005 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Original Article Braz. J. Vet. Parasitol., Jaboticabal, v. 26, n. 4, p. 433-438, oct.-dec. 2017 ISSN 0103-846X (Print) / ISSN 1984-2961 (Electronic) Doi: http://dx.doi.org/10.1590/S1984-29612017060 Litomosoides silvai (Nematoda: Onchocercidae) parasitizing Akodon montensis (Rodentia: Cricetidae) in the southern region of Brazil Litomosoides silvai (Nematoda: Onchocercidae) parasitando Akodon montensis (Rodentia: Cricetidae) na região Sul do Brasil Lucas Trevisan Gressler1; Felipe da Silva Krawczak2,3; Marcelo Knoff4; Silvia Gonzalez Monteiro1*; Marcelo Bahia Labruna2; -

Investigating Evolutionary Processes Using Ancient and Historical DNA of Rodent Species
Investigating evolutionary processes using ancient and historical DNA of rodent species Thesis submitted for the degree of Doctor of Philosophy (PhD) University of London Royal Holloway University of London Egham, Surrey TW20 OEX Selina Brace November 2010 1 Declaration I, Selina Brace, declare that this thesis and the work presented in it is entirely my own. Where I have consulted the work of others, it is always clearly stated. Selina Brace Ian Barnes 2 “Why should we look to the past? ……Because there is nowhere else to look.” James Burke 3 Abstract The Late Quaternary has been a period of significant change for terrestrial mammals, including episodes of extinction, population sub-division and colonisation. Studying this period provides a means to improve understanding of evolutionary mechanisms, and to determine processes that have led to current distributions. For large mammals, recent work has demonstrated the utility of ancient DNA in understanding demographic change and phylogenetic relationships, largely through well-preserved specimens from permafrost and deep cave deposits. In contrast, much less ancient DNA work has been conducted on small mammals. This project focuses on the development of ancient mitochondrial DNA datasets to explore the utility of rodent ancient DNA analysis. Two studies in Europe investigate population change over millennial timescales. Arctic collared lemming (Dicrostonyx torquatus) specimens are chronologically sampled from a single cave locality, Trou Al’Wesse (Belgian Ardennes). Two end Pleistocene population extinction-recolonisation events are identified and correspond temporally with - localised disappearance of the woolly mammoth (Mammuthus primigenius). A second study examines postglacial histories of European water voles (Arvicola), revealing two temporally distinct colonisation events in the UK. -

Rodentia, Cricetidae) and Their Hybrids
_??_1995 The Japan Mendel Society Cytologia 60: 93-102 , 1995 Chromosomal and Synaptonemal Complex Analysis of Robertsonian Polymorphisms in Akodon dolores and Akodon molinae (Rodentia, Cricetidae) and their Hybrids P. Wittouck, E. Pinna Senn, C. A. Sonez, M. C. Provensal, J. J. Polop and J. A. Lisanti Departamento de Ciencias Naturales. Universidad Nacional de Rio Cuarto . (5800). Rio Cuarto, Argentina Accepted March 9, 1995 The complex cricetid genus Akodon, with its five subgenera and more than 40 species (Apfelbaum and Reig 1989, Reig 1987) is characterized by several features that make it very interesting for cytogenetical research, such as intraspecific variation of the autosomes and sex chromosomes, the existence of XY fertile females in some species, and the presence of different species that show apparently identical karyotypes (Bianchi and Merani 1984, Bianchi et al. 1971, 1979a, b, 1989, Gallardo 1982, Maia and Langguth 1981, Reig 1987, Vitullo et al. 1986, and references therein). Two closely related species of the genus, Akodon dolores and A. molinae, share the G-band pattern of their chromosomal arms (Bianchi et al. 1979a) and produce fertile hybrids, at least in laboratory conditions (Merani et al. 1978, Roldan et al. 1984). Their populations present Robertsonian polymorphisms affecting one chromosome pair in A. molinae (Bianchi et al. 1973), and this one and several other pairs in A. dolores (Bianchi et al. 1979a). We report here karyological analyses on some populations assigned to A. dolores on morphological grounds and on hybrid specimens, and show that one of these populations corresponds karyotypically to A. molinae. Craniometric studies fail to differentiate between populations with "dolores" or "molinae" karyotype .