Table SI. the 333 Representative Protein Sequences of Nrs Were Used in the Present Study
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Alpha Actinin 4: an Intergral Component of Transcriptional
ALPHA ACTININ 4: AN INTERGRAL COMPONENT OF TRANSCRIPTIONAL PROGRAM REGULATED BY NUCLEAR HORMONE RECEPTORS By SIMRAN KHURANA Submitted in partial fulfillment of the requirements for the degree of doctor of philosophy Thesis Advisor: Dr. Hung-Ying Kao Department of Biochemistry CASE WESTERN RESERVE UNIVERSITY August, 2011 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of SIMRAN KHURANA ______________________________________________________ PhD candidate for the ________________________________degree *. Dr. David Samols (signed)_______________________________________________ (chair of the committee) Dr. Hung-Ying Kao ________________________________________________ Dr. Edward Stavnezer ________________________________________________ Dr. Leslie Bruggeman ________________________________________________ Dr. Colleen Croniger ________________________________________________ ________________________________________________ May 2011 (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. TABLE OF CONTENTS LIST OF TABLES vii LIST OF FIGURES viii ACKNOWLEDEMENTS xii LIST OF ABBREVIATIONS xiii ABSTRACT 1 CHAPTER 1: INTRODUCTION Family of Nuclear Receptors 3 Mechanism of transcriptional regulation by co-repressors and co-activators 8 Importance of LXXLL motif of co-activators in NR mediated transcription 12 Cyclic recruitment of co-regulators on the target promoters 15 Actin and actin related proteins (ABPs) in transcription -

Role of Nuclear Receptors in Central Nervous System Development and Associated Diseases
Role of Nuclear Receptors in Central Nervous System Development and Associated Diseases The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Olivares, Ana Maria, Oscar Andrés Moreno-Ramos, and Neena B. Haider. 2015. “Role of Nuclear Receptors in Central Nervous System Development and Associated Diseases.” Journal of Experimental Neuroscience 9 (Suppl 2): 93-121. doi:10.4137/JEN.S25480. http:// dx.doi.org/10.4137/JEN.S25480. Published Version doi:10.4137/JEN.S25480 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:27320246 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Journal name: Journal of Experimental Neuroscience Journal type: Review Year: 2015 Volume: 9(S2) Role of Nuclear Receptors in Central Nervous System Running head verso: Olivares et al Development and Associated Diseases Running head recto: Nuclear receptors development and associated diseases Supplementary Issue: Molecular and Cellular Mechanisms of Neurodegeneration Ana Maria Olivares1, Oscar Andrés Moreno-Ramos2 and Neena B. Haider1 1Department of Ophthalmology, Schepens Eye Research Institute, Massachusetts Eye and Ear, Harvard Medical School, Boston, MA, USA. 2Departamento de Ciencias Biológicas, Facultad de Ciencias, Universidad de los Andes, Bogotá, Colombia. ABSTRACT: The nuclear hormone receptor (NHR) superfamily is composed of a wide range of receptors involved in a myriad of important biological processes, including development, growth, metabolism, and maintenance. -

Anti-Testicular Receptor 2 (TR2) (Rabbit) Antibody - 100-401-E45
Anti-Testicular Receptor 2 (TR2) (Rabbit) Antibody - 100-401-E45 Code: 100-401-E45 Size: 100 µL Product Description: Anti-Testicular Receptor 2 (TR2) (Rabbit) Antibody - 100-401-E45 Concentration: Titrated value sufficient to run approximately 10 mini blots. PhysicalState: Liquid Label Unconjugated Host Rabbit Gene Name NR2C1 Species Reactivity mouse, human, rat Storage Condition Store vial at -20° C prior to opening. This product is stable at 4° C as an undiluted liquid. For extended storage, aliquot contents and freeze at -20° C or below. Avoid cycles of freezing and thawing. Dilute only prior to immediate use. Synonyms Nuclear receptor subfamily 2 group C member 1, Orphan nuclear receptor TR2, mTR2 Application Note Anti-Testicular Receptor 2 (Rabbit) antibody is suitable for use in Western Blots. Anti-Testicular Receptor 2 antibodies are specific for the ~64 kDa TR2 protein in Western blots of testes and nuclear extracts from MEL cell lines. Researchers should determine optimal titers for applications that are not stated below. Background TR2 antibody detects Testicular receptor 2 (TR2), which is a member of the orphan nuclear receptor family. It is widely expressed at a low level throughout the adult testis. TR2 represses transcription and binds DNA directly interacting with HDAC3 and HDAC4 via DNA-binding domains. TR2 has also been implicated in regulation of estrogen receptor activity in mammary glands. In addition, TR2 has recently been shown to form a heterodimer with TR4 that can bind to the direct repeat 6 element of the hepatitis B virus (HBV) enhancer II region thus suppressing HBV gene expression. -

Alternative Splicing in the Nuclear Receptor Superfamily Expands Gene Function to Refine Endo-Xenobiotic Metabolism S
Supplemental material to this article can be found at: http://dmd.aspetjournals.org/content/suppl/2020/01/24/dmd.119.089102.DC1 1521-009X/48/4/272–287$35.00 https://doi.org/10.1124/dmd.119.089102 DRUG METABOLISM AND DISPOSITION Drug Metab Dispos 48:272–287, April 2020 Copyright ª 2020 by The American Society for Pharmacology and Experimental Therapeutics Minireview Alternative Splicing in the Nuclear Receptor Superfamily Expands Gene Function to Refine Endo-Xenobiotic Metabolism s Andrew J. Annalora, Craig B. Marcus, and Patrick L. Iversen Department of Environmental and Molecular Toxicology, Oregon State University, Corvallis, Oregon (A.J.A., C.B.M., P.L.I.) and United States Army Research Institute for Infectious Disease, Frederick, Maryland (P.L.I.) Received August 16, 2019; accepted December 31, 2019 ABSTRACT Downloaded from The human genome encodes 48 nuclear receptor (NR) genes, whose Exon inclusion options are differentially distributed across NR translated products transform chemical signals from endo- subfamilies, suggesting group-specific conservation of resilient func- xenobiotics into pleotropic RNA transcriptional profiles that refine tionalities. A deeper understanding of this transcriptional plasticity drug metabolism. This review describes the remarkable diversifica- expands our understanding of how chemical signals are refined and tion of the 48 human NR genes, which are potentially processed into mediated by NR genes. This expanded view of the NR transcriptome over 1000 distinct mRNA transcripts by alternative splicing (AS). The informs new models of chemical toxicity, disease diagnostics, and dmd.aspetjournals.org average human NR expresses ∼21 transcripts per gene and is precision-based approaches to personalized medicine. -

MAGE-A11 Is Activated Through TFCP2/ZEB1 Binding Sites De-Methylation As Well As Histone Modification and Facilitates ESCC Tumor Growth
www.impactjournals.com/oncotarget/ Oncotarget, 2018, Vol. 9, (No. 3), pp: 3365-3378 Research Paper MAGE-A11 is activated through TFCP2/ZEB1 binding sites de-methylation as well as histone modification and facilitates ESCC tumor growth Shina Liu1,*, Fei Liu1,*, Weina Huang1, Lina Gu1, Lingjiao Meng1, Yingchao Ju1,2, Yunyan Wu1, Juan Li1, Lihua Liu1 and Meixiang Sang1,3 1Research Center, the Fourth Hospital of Hebei Medical University, Shijiazhuang 050011, P. R. China 2Animal Center, the Fourth Hospital of Hebei Medical University, Shijiazhuang 050011, P. R. China 3Tumor Research Institute, the Fourth Hospital of Hebei Medical University, Shijiazhuang 050011, P. R. China *These authors contributed equally to this work Correspondence to: Meixiang Sang, email: [email protected] Keywords: MAGE-A11; ESCC; DNA methylation; histone acetylation; histone methylation Received: September 30, 2017 Accepted: November 15, 2017 Published: December 05, 2017 Copyright: Liu et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Recently, we have reported that the product of Melanoma Antigens Genes (MAGE) family member MAGE-A11 is an independent poor prognostic marker for esophageal squamous cell carcinoma (ESCC). However, the reason how MAGE-A11 is activated in ESCC progression still remains unclear. In the current study, we demonstrated that DNA methylation and the subsequent histone posttranslational modifications play crucial roles in the regulation of MAGE-A11 in ESCC progression. We found that the methylation rate of TFCP2/ZEB1 binding site on MAGE-A11 promoter in ESCC tissues and cells is higher than the normal esophageal epithelial tissues and cells. -

Molecular Signatures Differentiate Immune States in Type 1 Diabetes Families
Page 1 of 65 Diabetes Molecular signatures differentiate immune states in Type 1 diabetes families Yi-Guang Chen1, Susanne M. Cabrera1, Shuang Jia1, Mary L. Kaldunski1, Joanna Kramer1, Sami Cheong2, Rhonda Geoffrey1, Mark F. Roethle1, Jeffrey E. Woodliff3, Carla J. Greenbaum4, Xujing Wang5, and Martin J. Hessner1 1The Max McGee National Research Center for Juvenile Diabetes, Children's Research Institute of Children's Hospital of Wisconsin, and Department of Pediatrics at the Medical College of Wisconsin Milwaukee, WI 53226, USA. 2The Department of Mathematical Sciences, University of Wisconsin-Milwaukee, Milwaukee, WI 53211, USA. 3Flow Cytometry & Cell Separation Facility, Bindley Bioscience Center, Purdue University, West Lafayette, IN 47907, USA. 4Diabetes Research Program, Benaroya Research Institute, Seattle, WA, 98101, USA. 5Systems Biology Center, the National Heart, Lung, and Blood Institute, the National Institutes of Health, Bethesda, MD 20824, USA. Corresponding author: Martin J. Hessner, Ph.D., The Department of Pediatrics, The Medical College of Wisconsin, Milwaukee, WI 53226, USA Tel: 011-1-414-955-4496; Fax: 011-1-414-955-6663; E-mail: [email protected]. Running title: Innate Inflammation in T1D Families Word count: 3999 Number of Tables: 1 Number of Figures: 7 1 For Peer Review Only Diabetes Publish Ahead of Print, published online April 23, 2014 Diabetes Page 2 of 65 ABSTRACT Mechanisms associated with Type 1 diabetes (T1D) development remain incompletely defined. Employing a sensitive array-based bioassay where patient plasma is used to induce transcriptional responses in healthy leukocytes, we previously reported disease-specific, partially IL-1 dependent, signatures associated with pre and recent onset (RO) T1D relative to unrelated healthy controls (uHC). -

Rs 000-000-260 AKT/Pkba Substrate 1.0 Mg/Ml 500 Μ
Catalog Number Display Name Concentration ValueSize Default Unit List price - Rs 000-000-260 AKT/PKBa Substrate 1.0 mg/mL 500 µg 24,434.00 000-000-264 NFKB p65 (Rel A) pS276 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-264NP NFKB p65 (Rel A) S276 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-266 p65 (Rel A) pS529 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-266NP p65 (Rel A) S529 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-383 DYKDDDDK (FLAG®) Peptide 1 mg/mL 1.0 mg 12,285.00 000-000-398 ATM S1981 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-400 ATM pS1981 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-401 AKT Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-402 Angiopoietin 2 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-403 Angiopoietin 1 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-404 Osteopontin Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-405 NOTCH 1 (intra) (Human specific) Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-407 NOTCH 1 (Human specific) Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-408 NOTCH 2 (Human specific) Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-410 ASK-1 phospho specific pS83 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-410NP ASK-1 non phospho specific S83 Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-433 Huntington pS421 Control Phospho Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-450 Huntington S421 Control Non-Phospho Peptide 1.0 mg/mL 50 µg 13,787.00 000-000-H47 Triple FLAG® Peptide 1.0 mg/ml 1.0 mg 12,285.00 000-000-K95 Beta Amyloid 40, Peptide 1.0 mg/mL 1.0 mg 1,06,334.00 000-000-K95S Beta Amyloid 40, Peptide 1.0 mg/mL 0.5 mg 27,164.00 000-000-M33 LL-37, Rhodamine -

Tepzz¥ 9 Z4a T
(19) TZZ¥ _ Z_T (11) EP 3 219 204 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 20.09.2017 Bulletin 2017/38 A01K 67/00 (2006.01) C07K 14/47 (2006.01) C07K 19/00 (2006.01) C12N 1/00 (2006.01) (2006.01) (2006.01) (21) Application number: 17165360.3 C12N 5/02 C12N 5/10 C12N 15/12 (2006.01) C12N 15/62 (2006.01) (2006.01) (2006.01) (22) Date of filing: 02.05.2005 C12N 15/63 C12P 21/02 (84) Designated Contracting States: (72) Inventors: AT BE BG CH CY CZ DE DK EE ES FI FR GB GR • PALLI, Subba, Reddy HU IE IS IT LI LT LU MC NL PL PT RO SE SI SK TR Lexington, KY 40504 (US) • KUMAR, Mohan, Basavaraju (30) Priority: 30.04.2004 US 567294 P Ottawa, Ontario K1H 6Z5 (CA) 13.09.2004 US 609424 P 29.04.2005 US 118855 (74) Representative: D Young & Co LLP 120 Holborn (62) Document number(s) of the earlier application(s) in London EC1N 2DY (GB) accordance with Art. 76 EPC: 15188829.4 / 3 000 317 Remarks: 05743351.8 / 1 744 619 This application was filed on 06-04-2017 as a divisional application to the application mentioned (71) Applicant: Intrexon Corporation under INID code 62. Blacksburg, VA 24060 (US) (54) MUTANTRECEPTORS ANDTHEIR USE INA NUCLEAR RECEPTOR-BASEDINDUCIBLE GENE EXPRESSION SYSTEM (57) This invention relates to the field of biotechnol- expression of a gene in a host cell for applications such ogy or genetic engineering. -

Nuclear Receptors in Metazoan Lineages: the Cross-Talk Between Evolution and Endocrine Disruption
Nuclear Receptors in Metazoan lineages: the cross -talk between Evolution and Endocrine Disruption Elza Sofia Silva Fonseca Tese de Doutoramento apresentada à Faculdade de Ciências da Universidade do Porto Biologia D 2020 Nuclear Receptors in Metazoan lineages: the cross-talk between Evolution and Endocrine Disruption D Elza Sofia Silva Foseca Doutoramento em Biologia Departamento de Biologia 2020 Orientador Doutor Luís Filipe Costa Castro, Professor Auxiliar, Faculdade de Ciências da Universidade do Porto, Centro Interdisciplinar de Investigação Marinha e Ambiental (CIIMAR) Coorientador Professor Doutor Miguel Alberto Fernandes Machado e Santos, Professor Auxiliar, Faculdade de Ciências da Universidade do Porto Centro Interdisciplinar de Investigação Marinha e Ambiental (CIIMAR) FCUP i Nuclear Receptors in Metazoan lineages: the cross-talk between Evolution and Endocrine Disruption This thesis was supported by FCT (ref: SFRH/BD/100262/2014), Norte2020 and FEDER (Coral – Sustainable Ocean Exploitation – Norte-01-0145-FEDER-000036 and EvoDis – Norte-01-0145-FEDER-031342). ii FCUP Nuclear Receptors in Metazoan lineages: the cross-talk between Evolution and Endocrine Disruption The present thesis is organized into seven chapters. Chapter 1 consists of a general introduction, providing an overview on Metazoa definition, and a review on the current knowledge of evolution and function of nuclear receptors and their role in endocrine disruption processes. Chapters 2, 3, 4 and 6 correspond to several projects developed during the doctoral programme presented here as independent articles, listed below (three articles published in peer reviewed international journals and one article in final preparation for submission). Chapter 5 was adapted from an article published in a peer reviewed international journal (listed below), in which I executed the methodology regarding the structural and functional analyses of rotifer RXR and I contributed to the writing of the sections referring to these analyses (Material and Methods, Results and Discussion). -

A SILAC-Based DNA Protein Interaction Screen That Identifies Candidate Binding Proteins to Functional DNA Elements
Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Methods A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements Gerhard Mittler,1,2,4 Falk Butter,3 and Matthias Mann3,5 1Center for Experimental Bioinformatics, University of Southern Denmark, DK-5230 Odense M, Denmark; 2BIOSS—Center of Biological Signalling Studies, Albert-Ludwigs-University Freiburg, D-79104 Freiburg, Germany; 3Department of Proteomics and Signal Transduction, Max-Planck-Institute for Biochemistry, D-82152 Martinsried, Germany Determining the underlying logic that governs the networks of gene expression in higher eukaryotes is an important task in the post-genome era. Sequence-specific transcription factors (TFs) that can read the genetic regulatory information and proteins that interpret the information provided by CpG methylation are crucial components of the system that controls the transcription of protein-coding genes by RNA polymerase II. We have previously described Stable Isotope Labeling by Amino acids in Cell culture (SILAC) for the quantitative comparison of proteomes and the determination of protein– protein interactions. Here, we report a generic and scalable strategy to uncover such DNA protein interactions by SILAC that uses a fast and simple one-step affinity capture of TFs from crude nuclear extracts. Employing mutated or non- methylated control oligonucleotides, specific TFs binding to their wild-type or methyl-CpG bait are distinguished from the vast excess of copurifying background proteins by their peptide isotope ratios that are determined by mass spec- trometry. Our proof of principle screen identifies several proteins that have not been previously reported to be present on the fully methylated CpG island upstream of the human metastasis associated 1 family, member 2 gene promoter. -

Translational Alterations of Retinoid Receptors, Their Binding Partners and The
Translational alterations of retinoid receptors, their binding partners and the retinoid signalling pathway in schizophrenia brain and blood Shan-Yuan Tsai Submitted in total fulfilment of the requirements of the degree of Doctor of Philosophy School of Psychiatry Faculty of Medicine August 2018 Thesis/Dissertation Sheet Australia's Global University Surname/Family Name Tsai Given Name/s Shan-Yuan Abbreviation for degree as give in the University calendar PhD Faculty Faulty of Medicine School School of Psychiatry Translational alt�ations of retinoid receptors,their binding partners and the Thesis Tille retinoid signalling pathway in schizophrenia brain and blood Abstract350 words maximum: (PLEASE TYPE) Schizophrenia is a debilitating mentalillness characterised by positive, negative and cognitive symptoms. Epidemiological studies implicate vitamin D in the aetiologyof schizophrenia and protein studies show altered circulating levels of the ligand vitamin D and implicate relinoids in schizophrenia. However, the gene expressions of vitamin D and rebnoid receptors have not beenext ensively studied. In parallel, there is increasing interest in the role ofneuroinflammation in the aetiology and progressionof schizophrenia and both vitamin D and retJnoids (vitamin A andits derivatives) have known anti-inflammatory actions. My thesis aims to measure expressionof molecules involved in vitamin D andretino id signalling and determine 1heinfluence of elevated inflammatory markers on these molecules. My findings are obtained from two cohoris: a post -
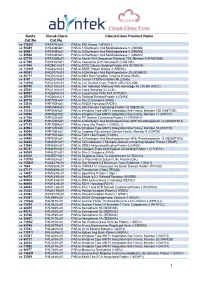
1 Santa Cat.No Cloud-Clone Cat.No. Cloud-Clone Product Name
Santa Cloud -Clone Cloud -Clone Product Name Cat.No Cat.No. sc -130253 PAS477Hu01 PAB to RIO Kinase 1 (RIOK1) sc -50485 PAS446Ra01 PAB to A Disintegrin And Metalloprotease 5 (ADAM5) sc -50487 PAS445Ra01 PAB to A Disintegrin And Metalloprotease 6 (ADAM6) sc -25588 PAS444Ra01 PAB to A Disintegrin And Metalloprotease 1 (ADAM1) sc -87719 PAR758Mu01 PAB to Family With Sequence Similarity 135, Member B (FAM135B) sc -67296 PAR493Hu01 PAB to Coenzyme Q10 Homolog B (COQ10B) sc -67048 PAQ981Hu01 PAB to S100 Calcium Binding Protein A15 (S100A15) sc -130269 PAQ342Hu01 PAB to SRSF Protein Kinase 3 (SRPK3) sc -98582 PAQ207Hu01 PAB to A Disintegrin And Metalloprotease 20 (ADAM20) sc -20711 PAQ164Hu01 PAB to BMX Non Receptor Tyrosine Kinase (BMX) sc -9147 PAQ127Hu01 PAB to Cluster Of Differentiation 8b (CD8b) sc -130184 PAQ124Hu01 PAB to Cell Division Cycle Protein 25B (CDC25B) sc -98790 PAQ118Hu01 PAB to Cell Adhesion Molecule With Homology To L1CAM (CHL1) sc -25361 PAQ116Hu01 PAB to Clock Homolog (CLOCK) sc -98937 PAQ089Hu01 PAB to Cytochrome P450 3A7 (CYP3A7) sc -25519 PAQ088Hu01 PAB to Dickkopf Related Protein 4 (DKK4) sc -28778 PAP797Hu01 PAB to Pim-2 Oncogene (PIM2) sc -33626 PAP750Hu01 PAB to RAD51 Homolog (RAD51) sc -8333 PAP694Mu01 PAB to SH2 Domain Containing Protein 1A (SH2D1A) sc -25524 PAP553Hu01 PAB to Wingless Type MMTV Integration Site Family, Member 10B (WNT10B) sc -50360 PAP552Hu01 PAB to Wingless Type MMTV Integration Site Family, Member 11 (WNT11) sc -87368 PAP332Hu01 PAB to PR Domain Containing Protein 14 (PRDM14) sc -25583 PAP226Hu01