DNA from Mollusc Shell: a Valuable and Underutilised Substrate for Genetic Analyses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Impact Damage and Repair in Shells of the Limpet Patella Vulgata David Taylor
© 2016. Published by The Company of Biologists Ltd | Journal of Experimental Biology (2016) 219, 3927-3935 doi:10.1242/jeb.149880 RESEARCH ARTICLE Impact damage and repair in shells of the limpet Patella vulgata David Taylor ABSTRACT can be a major cause of lethal damage. Shanks and Wright (1986) Experiments and observations were carried out to investigate the demonstrated the destructive power of wave-borne missiles by response of the Patella vulgata limpet shell to impact. Dropped-weight setting up targets to record impacts in a given area. They studied four impact tests created damage that usually took the form of a hole in the limpet species, finding that they were much more likely to be lost in shell’s apex. Similar damage was found to occur naturally, a location where there were many movable rocks and pebbles presumably as a result of stones propelled by the sea during compared with a location consisting largely of solid rock mass. storms. Apex holes were usually fatal, but small holes were Examining the populations over a one year period, and making a sometimes repaired, and the repaired shell was as strong as the number of assumptions about size distributions and growth rates, original, undamaged shell. The impact strength (energy to failure) of they estimated that 47% of shells were destroyed in the former shells tested in situ was found to be 3.4-times higher than that of location, compared with 7% in the latter (Shanks and Wright, 1986). empty shells found on the beach. Surprisingly, strength was not Another study (Cadée, 1999) reported that impact damage by ice affected by removing the shell from its home location, or by removing blocks and stones was the major cause of damage to the limpet the limpet from the shell and allowing the shell to dry out. -
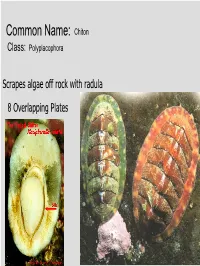
Common Name: Chiton Class: Polyplacophora
Common Name: Chiton Class: Polyplacophora Scrapes algae off rock with radula 8 Overlapping Plates Phylum? Mollusca Class? Gastropoda Common name? Brown sea hare Class? Scaphopoda Common name? Tooth shell or tusk shell Mud Tentacle Foot Class? Gastropoda Common name? Limpet Phylum? Mollusca Class? Bivalvia Class? Gastropoda Common name? Brown sea hare Phylum? Mollusca Class? Gastropoda Common name? Nudibranch Class? Cephalopoda Cuttlefish Octopus Squid Nautilus Phylum? Mollusca Class? Gastropoda Most Bivalves are Filter Feeders A B E D C • A: Mantle • B: Gill • C: Mantle • D: Foot • E: Posterior adductor muscle I.D. Green: Foot I.D. Red Gills Three Body Regions 1. Head – Foot 2. Visceral Mass 3. Mantle A B C D • A: Radula • B: Mantle • C: Mouth • D: Foot What are these? Snail Radulas Dorsal HingeA Growth line UmboB (Anterior) Ventral ByssalC threads Mussel – View of Outer Shell • A: Hinge • B: Umbo • C: Byssal threads Internal Anatomy of the Bay Mussel A B C D • A: Labial palps • B: Mantle • C: Foot • D: Byssal threads NacreousB layer Posterior adductorC PeriostracumA muscle SiphonD Mantle Byssal threads E Internal Anatomy of the Bay Mussel • A: Periostracum • B: Nacreous layer • C: Posterior adductor muscle • D: Siphon • E: Mantle Byssal gland Mantle Gill Foot Labial palp Mantle Byssal threads Gill Byssal gland Mantle Foot Incurrent siphon Byssal Labial palp threads C D B A E • A: Foot • B: Gills • C: Posterior adductor muscle • D: Excurrent siphon • E: Incurrent siphon Heart G F H E D A B C • A: Foot • B: Gills • C: Mantle • D: Excurrent siphon • E: Incurrent siphon • F: Posterior adductor muscle • G: Labial palps • H: Anterior adductor muscle Siphon or 1. -

Silicified Eocene Molluscs from the Lower Murchison District, Southern Carnarvon Basin, Western Australia
[<ecords o{ the Western A uslralian Museum 24: 217--246 (2008). Silicified Eocene molluscs from the Lower Murchison district, Southern Carnarvon Basin, Western Australia Thomas A. Darragh1 and George W. Kendrick2.3 I Department of Invertebrate Palaeontology, Museum Victoria, 1'.0. Box 666, Melbourne, Victoria 3001, Australia. Email: tdarragh(il.Illuseum.vic.gov.au :' Department of Earth and Planetary Sciences, Western Australian Museum, Locked Bag 49, Welshpool D.C., Western Australia 6986, Australia. 1 School of Earth and Ceographical Sciences, The University of Western Australia, 35 Stirling Highway, Crawlev, Western Australia 6009, Australia. Abstract - Silicified Middle to Late Eocene shallow water sandstones outcropping in the Lower Murchison District near Kalbarri township contain a silicified fossil fauna including foraminifera, sponges, bryozoans, solitary corals, brachiopods, echinoids and molluscs. The known molluscan fauna consists of 51 species, comprising 2 cephalopods, 14 bivalves, 1 scaphopod and 34 gastropods. Of these taxa three are newly described, Cerithium lvilya, Zeacolpus bartol1i, and Lyria lamellatoplicata. 25 of these molluscs are identical to or closely comparable with taxa from the southern Australian Eocene. The occurrence of this fauna extends the Southeast Australian Province during the Eocene from southwest Western Australia along the west coast north to at least 27° present day south latitude; consequently the province is here renamed the Southern Australian Province. Keywords: siliceous fossils, Eocene, Kalbarri, molluscs, new taxa, Carnarvon Basin, biogeography, Southern Australian Province. INTRODUCTION The source deposit, a pallid to ferruginous silicified Eocene marine molluscan assemblages from sandstone, forms a weakly defined, low breakaway coastal sedimentary basins in southern Australia trending N-S and sloping gently westward. -
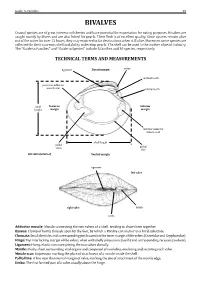
Field Identification Guide to the Living Marine Resources In
Guide to Families 29 BIVALVES Coastal species are of great interest to fisheries and have potential for exportation for eating purposes. Bivalves are caught mainly by divers and are also fished for pearls. Their flesh is of excellent quality. Since oysters remain alive out of the water for over 12 hours, they may exported to far destinations when still alive. Moreover, some species are collected for their nacreous shell and ability to develop pearls. The shell can be used in the mother of pearl industry. The “Guide to Families’’ andTECHNICAL ‘‘Guide to Species’’ TERMS include 5AND families MEASUREMENTS and 10 species, respectively. ligament Dorsal margin umbo posterior adductor cardinal tooth muscle scar lateral tooth Posterior Anterior margin margin shell height anterior adductor muscle scar pallial sinus pallial shell length line left valve (interior) Ventral margin ligament left valve right valve lunule umbo Adductor muscle: Byssus: Chomata: Muscle connecting the two valves of a shell, tending to draw them together. Hinge: Clump of horny threads spun by the foot, by which a Bivalve can anchor to a hard substrate. Ligament: Small denticles and corresponding pits located on the inner margin of the valves (Ostreidae and Gryphaeidae). Mantle: Top interlocking margin of the valves, often with shelly projections (teeth) and corresponding recesses (sockets). Muscle scar: Horny, elastic structure joining the two valves dorsally. Pallial line: Fleshy sheet surrounding vital organs and composed of two lobes, one lining and secreting each valve. Umbo: Impression marking the place of attachment of a muscle inside the shell. A line near the internal margin of valve, marking the site of attachment of the mantle edge. -

Clam Dissection Guideline
Clam Dissection Guideline BACKGROUND: Clams are bivalves, meaning that they have shells consisting of two halves, or valves. The valves are joined at the top, and the adductor muscles on each side hold the shell closed. If the adductor muscles are relaxed, the shell is pulled open by ligaments located on each side of the umbo. The clam's foot is used to dig down into the sand, and a pair of long incurrent and excurrent siphons that extrude from the clam's mantle out the side of the shell reach up to the water above (only the exit points for the siphons are shown). Clams are filter feeders. Water and food particles are drawn in through one siphon to the gills where tiny, hair-like cilia move the water, and the food is caught in mucus on the gills. From there, the food-mucus mixture is transported along a groove to the palps (mouth flaps) which push it into the clam's mouth. The second siphon carries away the water. The gills also draw oxygen from the water flow. The mantle, a thin membrane surrounding the body of the clam, secretes the shell. The oldest part of the clam shell is the umbo, and it is from the hinge area that the clam extends as it grows. I. Purpose: The purpose of this lab is to identify the internal and external structures of a mollusk by dissecting a clam. II. Materials: 2 pairs of safety goggles 1 paper towel 2 pairs of gloves 1 pair of scissors 1 preserved clam 2 pairs of forceps 1 dissecting tray 2 probes III. -

Os Nomes Galegos Dos Moluscos
A Chave Os nomes galegos dos moluscos 2017 Citación recomendada / Recommended citation: A Chave (2017): Nomes galegos dos moluscos recomendados pola Chave. http://www.achave.gal/wp-content/uploads/achave_osnomesgalegosdos_moluscos.pdf 1 Notas introdutorias O que contén este documento Neste documento fornécense denominacións para as especies de moluscos galegos (e) ou europeos, e tamén para algunhas das especies exóticas máis coñecidas (xeralmente no ámbito divulgativo, por causa do seu interese científico ou económico, ou por seren moi comúns noutras áreas xeográficas). En total, achéganse nomes galegos para 534 especies de moluscos. A estrutura En primeiro lugar preséntase unha clasificación taxonómica que considera as clases, ordes, superfamilias e familias de moluscos. Aquí apúntase, de maneira xeral, os nomes dos moluscos que hai en cada familia. A seguir vén o corpo do documento, onde se indica, especie por especie, alén do nome científico, os nomes galegos e ingleses de cada molusco (nalgún caso, tamén, o nome xenérico para un grupo deles). Ao final inclúese unha listaxe de referencias bibliográficas que foron utilizadas para a elaboración do presente documento. Nalgunhas desas referencias recolléronse ou propuxéronse nomes galegos para os moluscos, quer xenéricos quer específicos. Outras referencias achegan nomes para os moluscos noutras linguas, que tamén foron tidos en conta. Alén diso, inclúense algunhas fontes básicas a respecto da metodoloxía e dos criterios terminolóxicos empregados. 2 Tratamento terminolóxico De modo moi resumido, traballouse nas seguintes liñas e cos seguintes criterios: En primeiro lugar, aprofundouse no acervo lingüístico galego. A respecto dos nomes dos moluscos, a lingua galega é riquísima e dispomos dunha chea de nomes, tanto específicos (que designan un único animal) como xenéricos (que designan varios animais parecidos). -

Freshwater Mussels of Maritime Canada: a Flashcard Guide
Freshwater Mussels of Maritime Canada: A Flashcard Guide In Wolastoqey, Mi’kmaw, French and English UMBO DORSAL MARGIN ANTERIOR POSTERIOR MARGIN MARGIN VENTRAL MARGIN Donald F. McAlpine, Mary C. Sollows, Jacqueline B. Madill and André L. Martel ISBN 978-0-919326-80-4 All photos copyright the Canadian Museum of Nature Acknowledgements: Funding for this publication provided by the Department of Fisheries and Oceans and the New Brunswick Museum. Special thanks to Ree Brennin Houston, Department of Fisheries and Oceans; Anne Hamilton, Brent Suttie, New Brunswick Archaeological Services Branch, and indigenous language translators Allan Tremblay (Wolastoqiyk), George Paul, Howard Augustine, and Karen Narvey (Mi’kmaw). Citation: McAlpine, D.F., M.C. Sollows, J. B. Madill, and A. L. Martel. 2018. Freshwater Mussels of Maritime Canada: A Flashcard Guide in Wolastoqey, Mi’kmaw, French and English. New Brunswick Museum, Saint John, New Brunswick, and Canadian Museum of Nature, Ottawa, Canada. Use in conjunction with Martel, A. L.,D.F. McAlpine, J. Madill, D. Sabine, A. Paquet, M. Pulsifer and M. Elderkin. 2010. Pp. 551-598. Freshwater Mussels (Bivalvia: Margaritiferidae, Unionidae) of the Atlantic Maritime Ecozone. In D.F. McAlpine and I.M. Smith (eds.). Assessment of Species Diversity in the Atlantic Maritime Ecozone. NRC Research Press, National Research Council of Canada, Ottawa, ON. 785 pp. Nedeau, E.J., M.A. McCollough, and B.I. Swartz. 2000. Freshwater Mussels of Maine. Maine Department of Inland Fisheries and Wildlife, Augusta, ME, 118 pp. -

Marine Boring Bivalve Mollusks from Isla Margarita, Venezuela
ISSN 0738-9388 247 Volume: 49 THE FESTIVUS ISSUE 3 Marine boring bivalve mollusks from Isla Margarita, Venezuela Marcel Velásquez 1 1 Museum National d’Histoire Naturelle, Sorbonne Universites, 43 Rue Cuvier, F-75231 Paris, France; [email protected] Paul Valentich-Scott 2 2 Santa Barbara Museum of Natural History, Santa Barbara, California, 93105, USA; [email protected] Juan Carlos Capelo 3 3 Estación de Investigaciones Marinas de Margarita. Fundación La Salle de Ciencias Naturales. Apartado 144 Porlama,. Isla de Margarita, Venezuela. ABSTRACT Marine endolithic and wood-boring bivalve mollusks living in rocks, corals, wood, and shells were surveyed on the Caribbean coast of Venezuela at Isla Margarita between 2004 and 2008. These surveys were supplemented with boring mollusk data from malacological collections in Venezuelan museums. A total of 571 individuals, corresponding to 3 orders, 4 families, 15 genera, and 20 species were identified and analyzed. The species with the widest distribution were: Leiosolenus aristatus which was found in 14 of the 24 localities, followed by Leiosolenus bisulcatus and Choristodon robustus, found in eight and six localities, respectively. The remaining species had low densities in the region, being collected in only one to four of the localities sampled. The total number of species reported here represents 68% of the boring mollusks that have been documented in Venezuelan coastal waters. This study represents the first work focused exclusively on the examination of the cryptofaunal mollusks of Isla Margarita, Venezuela. KEY WORDS Shipworms, cryptofauna, Teredinidae, Pholadidae, Gastrochaenidae, Mytilidae, Petricolidae, Margarita Island, Isla Margarita Venezuela, boring bivalves, endolithic. INTRODUCTION The lithophagans (Mytilidae) are among the Bivalve mollusks from a range of families have more recognized boring mollusks. -

Pleistocene Molluscs from the Namaqualand Coast
ANNALS OF THE SOUTH AFRICAN MUSEUM ANNALE VAN DIE SUID-AFRIKAANSE MUSEUM Volume 52 Band July 1969 Julie Part 9 Dee! PLEISTOCENE MOLLUSCS FROM THE NAMAQUALAND COAST By A.J.CARRINGTON & B.F.KENSLEY are issued in parts at irregular intervals as material becomes available Obtainable from the South African Museum, P.O. Box 61, Cape Town word uitgegee in dele opongereelde tye na beskikbaarheid van stof OUT OF PRINT/UIT nRUK I, 2(1, 3, 5, 7-8), 3(1-2, 5, t.-p.i.), 5(2, 5, 7-9), 6(1, t.-p.i.), 7(1, 3), 8, 9(1-2), 10(1-3), 11(1-2, 7, t.-p.i.), 21, 24(2), 27, 31(1-3), 38, 44(4)· Price of this part/Prys van hierdie deel Rg.oo Trustees of the South African Museum © 1969 Printed in South Africa by In Suid-Afrika gedruk deur The Rustica Press, Pty., Ltd. Die Rustica-pers, Edms., Bpk. Court Road, Wynberg, Cape Courtweg, Wynberg, Kaap By A. ]. CARRINGTON & B. F. KENSLEY South African Museum, Cape Town (With plates 18 to 29 and I I figures) PAGE Introduction 189 Succession 190 Systematic discussion. 191 Acknowledgements 222 Summary. 222 References 223 INTRODUCTION In the course of an examination of the Tertiary to Recent sediments of the Namaqualand coast, being carried out by one of the authors (A.].C.), a collection of fossil molluscs was assembled from the Pleistocene horizons encountered in the area. The purpose of this paper is to introduce and describe some twenty species from this collection, including forms new to the South Mrican palaeontological literature. -

Missouri's Freshwater Mussels
Missouri mussel invaders Two exotic freshwater mussels, the Asian clam (Corbicula and can reproduce at a much faster rate than native mussels. MISSOURI’S fluminea) and the zebra mussel (Dreissena polymorpha), have Zebra mussels attach to any solid surface, including industrial found their way to Missouri. The Asian clam was introduced pipes, native mussels and snails and other zebra mussels. They into the western U.S. from Asia in the 1930s and quickly spread form dense clumps that suffocate and kill native mussels by eastward. Since 1968 it has spread rapidly throughout Missouri restricting feeding, breathing and other life functions. Freshwater and is most abundant in streams south of the Missouri River. In You can help stop the spread of these mussels by not moving the mid-1980s, zebra mussels hitched a ride in the ballast waters bait or boat well water from one stream to another; dump and of freighter ships traveling from Asia to the Great Lakes. They drain on the ground before leaving. Check all surfaces of your have rapidly moved into the Mississippi River basin and boat and trailer for zebra mussels and destroy them, along with westward to Oklahoma. vegetation caught on the boat or trailer. Wash with hot (104˚F) Asian clam and zebra mussel larvae have an advantage here water at a carwash and allow all surfaces to dry in the sun for at because they don’t require a fish host to reach a juvenile stage least five days before boating again. MusselsMusselsSue Bruenderman, Janet Sternburg and Chris Barnhart Zebra mussels attached to a native mussel JIM RATHERT ZEBRA CHRIS BARNHART ASIAN CLAM MUSSEL Shells are very common statewide in rivers, ponds and reservoirs A female can produce more than a million larvae at one time, and are often found on banks and gravel bars. -

Nabs 2004 Final
CURRENT AND SELECTED BIBLIOGRAPHIES ON BENTHIC BIOLOGY 2004 Published August, 2005 North American Benthological Society 2 FOREWORD “Current and Selected Bibliographies on Benthic Biology” is published annu- ally for the members of the North American Benthological Society, and summarizes titles of articles published during the previous year. Pertinent titles prior to that year are also included if they have not been cited in previous reviews. I wish to thank each of the members of the NABS Literature Review Committee for providing bibliographic information for the 2004 NABS BIBLIOGRAPHY. I would also like to thank Elizabeth Wohlgemuth, INHS Librarian, and library assis- tants Anna FitzSimmons, Jessica Beverly, and Elizabeth Day, for their assistance in putting the 2004 bibliography together. Membership in the North American Benthological Society may be obtained by contacting Ms. Lucinda B. Johnson, Natural Resources Research Institute, Uni- versity of Minnesota, 5013 Miller Trunk Highway, Duluth, MN 55811. Phone: 218/720-4251. email:[email protected]. Dr. Donald W. Webb, Editor NABS Bibliography Illinois Natural History Survey Center for Biodiversity 607 East Peabody Drive Champaign, IL 61820 217/333-6846 e-mail: [email protected] 3 CONTENTS PERIPHYTON: Christine L. Weilhoefer, Environmental Science and Resources, Portland State University, Portland, O97207.................................5 ANNELIDA (Oligochaeta, etc.): Mark J. Wetzel, Center for Biodiversity, Illinois Natural History Survey, 607 East Peabody Drive, Champaign, IL 61820.................................................................................................................6 ANNELIDA (Hirudinea): Donald J. Klemm, Ecosystems Research Branch (MS-642), Ecological Exposure Research Division, National Exposure Re- search Laboratory, Office of Research & Development, U.S. Environmental Protection Agency, 26 W. Martin Luther King Dr., Cincinnati, OH 45268- 0001 and William E. -

TREATISE ONLINE Number 48
TREATISE ONLINE Number 48 Part N, Revised, Volume 1, Chapter 31: Illustrated Glossary of the Bivalvia Joseph G. Carter, Peter J. Harries, Nikolaus Malchus, André F. Sartori, Laurie C. Anderson, Rüdiger Bieler, Arthur E. Bogan, Eugene V. Coan, John C. W. Cope, Simon M. Cragg, José R. García-March, Jørgen Hylleberg, Patricia Kelley, Karl Kleemann, Jiří Kříž, Christopher McRoberts, Paula M. Mikkelsen, John Pojeta, Jr., Peter W. Skelton, Ilya Tëmkin, Thomas Yancey, and Alexandra Zieritz 2012 Lawrence, Kansas, USA ISSN 2153-4012 (online) paleo.ku.edu/treatiseonline PART N, REVISED, VOLUME 1, CHAPTER 31: ILLUSTRATED GLOSSARY OF THE BIVALVIA JOSEPH G. CARTER,1 PETER J. HARRIES,2 NIKOLAUS MALCHUS,3 ANDRÉ F. SARTORI,4 LAURIE C. ANDERSON,5 RÜDIGER BIELER,6 ARTHUR E. BOGAN,7 EUGENE V. COAN,8 JOHN C. W. COPE,9 SIMON M. CRAgg,10 JOSÉ R. GARCÍA-MARCH,11 JØRGEN HYLLEBERG,12 PATRICIA KELLEY,13 KARL KLEEMAnn,14 JIřÍ KřÍž,15 CHRISTOPHER MCROBERTS,16 PAULA M. MIKKELSEN,17 JOHN POJETA, JR.,18 PETER W. SKELTON,19 ILYA TËMKIN,20 THOMAS YAncEY,21 and ALEXANDRA ZIERITZ22 [1University of North Carolina, Chapel Hill, USA, [email protected]; 2University of South Florida, Tampa, USA, [email protected], [email protected]; 3Institut Català de Paleontologia (ICP), Catalunya, Spain, [email protected], [email protected]; 4Field Museum of Natural History, Chicago, USA, [email protected]; 5South Dakota School of Mines and Technology, Rapid City, [email protected]; 6Field Museum of Natural History, Chicago, USA, [email protected]; 7North