Construction of Prognostic Microrna Signature for Human Invasive Breast Cancer by Integrated Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dissert 1 Title
GENETIC AND BIOCHEMICAL ANALYSIS OF ESSENTIAL ENZYMES IN TRIACYLGLYCEROL SYNTHESIS IN ARABIDOPSIS By KIMBERLY LYNN COTTON A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY WASHINGTON STATE UNIVERSITY Molecular Plant Sciences DECEMBER 2015 © Copyright by KIMBERLY LYNN COTTON, 2015 All Rights Reserved To the Faculty of Washington State University: The members of the Committee appointed to examine the dissertation of KIMBERLY LYNN COTTON find it satisfactory and recommend that it be accepted. ____________________________________________ John A. Browse, Ph.D., Chair _____________________________________________ Michael M. Neff, Ph.D. _____________________________________________ Thomas W. Okita, Ph.D. _____________________________________________ John J. Wyrick, Ph.D. ii ACKNOWLEDGMENTS To my PI John Browse, thank you for your support, guidance and patience over the years. To my committee, John Browse, Michael Neff, Thomas Okita, and John Wyrick, thank you for your insightful discussions and suggestions. To my collaborators Jay Shockey, Anushobha Regmi, Neil Adhikari, John Browse, and Phil Bates, thanks for the great work and great times. To Browse Lab members past and present, thanks for the help, guidance and wonderful memories. To the undergraduate students who have helped me over the years, thank you for all your help in making this project possible; and with a special thank you to Arlene, Tesa, Breana, and “no-longer-undergrads” David and Barbara Cotton for their particular dedication to this project. And finally, a huge thank you to my family and friends who have loved and supported me throughout this venture. Go Cougs! iii GENETIC AND BIOCHEMICAL ANALYSIS OF ESSENTIAL ENZYMES IN TRIACYLGLYCEROL SYNTHESIS IN ARABIDOPSIS Abstract by Kimberly Lynn Cotton, Ph.D. -

Anti-Inflammatory Role of Curcumin in LPS Treated A549 Cells at Global Proteome Level and on Mycobacterial Infection
Anti-inflammatory Role of Curcumin in LPS Treated A549 cells at Global Proteome level and on Mycobacterial infection. Suchita Singh1,+, Rakesh Arya2,3,+, Rhishikesh R Bargaje1, Mrinal Kumar Das2,4, Subia Akram2, Hossain Md. Faruquee2,5, Rajendra Kumar Behera3, Ranjan Kumar Nanda2,*, Anurag Agrawal1 1Center of Excellence for Translational Research in Asthma and Lung Disease, CSIR- Institute of Genomics and Integrative Biology, New Delhi, 110025, India. 2Translational Health Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India. 3School of Life Sciences, Sambalpur University, Jyoti Vihar, Sambalpur, Orissa, 768019, India. 4Department of Respiratory Sciences, #211, Maurice Shock Building, University of Leicester, LE1 9HN 5Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia- 7003, Bangladesh. +Contributed equally for this work. S-1 70 G1 S 60 G2/M 50 40 30 % of cells 20 10 0 CURI LPSI LPSCUR Figure S1: Effect of curcumin and/or LPS treatment on A549 cell viability A549 cells were treated with curcumin (10 µM) and/or LPS or 1 µg/ml for the indicated times and after fixation were stained with propidium iodide and Annexin V-FITC. The DNA contents were determined by flow cytometry to calculate percentage of cells present in each phase of the cell cycle (G1, S and G2/M) using Flowing analysis software. S-2 Figure S2: Total proteins identified in all the three experiments and their distribution betwee curcumin and/or LPS treated conditions. The proteins showing differential expressions (log2 fold change≥2) in these experiments were presented in the venn diagram and certain number of proteins are common in all three experiments. -
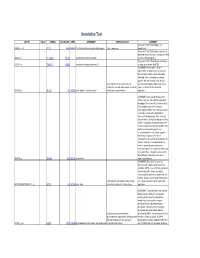
Supplementary Table 5. Functional Annotation of the Largest Gene Cluster(221 Element)
Annotation Tool AFFYID VALUE SYMBOL LOCUSLINK OMIM GENENAME GENEONTOLOGY SUMMARY [Proteome FUNCTION:] Expressed 203054_s_at TCTA 6988 600690 T-cell leukemia translocation altered gene tumor suppressor ubiquitously [Proteome FUNCTION:] May be involved in protein-protein interactions; contains five WD 44563_at FLJ10385 55135 hypothetical protein FLJ10385 domains (WD-40 repeats) [Proteome FUNCTION:] Weakly similarity to 212261_at TNRC15 26058 trinucleotide repeat containing 15 a region of rat nestin (Rn.9701) [SUMMARY:] Actin alpha 1 which is expressed in skeletal muscle is one of six different actin isoforms which have been identified. Actins are highly conserved proteins that are involved in cell motility, actin filament; motor activity; muscle structure and integrity. Alpha actins are a contraction; muscle development; structural major constituent of the contractile 203872_at ACTA1 58 102610 actin, alpha 1, skeletal muscle constituent of cytoskeleton apparatus. [SUMMARY:] Annexin VIII belong to the family of Ca (2+) dependent phospholipid binding proteins (annexins), and has a high 56% identity to annexin V (vascular anticoagulant-alpha). It was initially isolated as 2.2 kb vascular anticoagulant-beta transcript from human placenta, a Ca (2+) dependent phospholipid binding protein that inhibits coagulation and phospholipase A2 activity. However, the fact that annexin VIII is neither an extracellular protein nor associated with the cell surface suggests that it may not play a role in blood coagulation in vivo and its physiological role remains unknown. It is expressed at low levels in human placenta and shows restricted expression in lung endothelia, skin, liver, and kidney. The gene is also found to be selectively overexpressed in acute 203074_at ANXA8 244 602396 annexin A8 myelocytic leukemia. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

Mirnas Documented to Play a Role in Hematopoietic Cell Lineage. Our
Table S1: miRNAs documented to play a role in hematopoietic cell lineage. Our review of the literature summarizing miRNAs known to be involved in the development and proliferation of the hematopoietic lineage cells. miRNA Expression/function/target/regulator References miR-150 Elevated during developmental stages of B and T cell maturation. 16-19 Controls B cell differentiation, present in mature, resting T and B cells but decreased upon activation of naïve T or B cells. Plays a role in establishing lymphocyte identity. Very little is known about function in T cells. Regulators: Foxp3 Target: C-Myb miR-146a/b Upregulated in macropgahe inflammatory response. Differentially 17, 20 upregulated in murine Th1 subset but abolished in Th2 subset. Upregulated in response to TCRs stimulation, as well as by IL-1 and TNF. Highly expressed in murine T-regs and could play a role in establishing lymphocyte identity. Modulates activation induced cell death in activated T cells. Negative regulator of TLR and cytokine signaling pathway. Endotoxin tolerance. Antiviral role. Targets: IRAK1, IRAK2, TRAF6, FAF1 miR-16-1 Promote apoptosis by targeting Bcl2 expression, act as tumor 22 cluster suppressor RNAs. May block differentiation of later stage hematopoietic progenitor cells to mature cells. Downregulated in CLL. Target: BCL2. miR-155 Regulator of T and B cell maturation and innate immune response. 23-29 Expressed in primary mediastinal B-cell lymphoma, T and B cells, macrophages and DCs. Upregulated during B cell activation. Involved in T cell differentiation and indicated as a positive regulator of cytokine production. Activated by stimulating TLR3 and INFab receptors in bone derived macrophages (regulation of antimicrobial defense). -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

(12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur Et Al
US 20150240226A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur et al. (43) Pub. Date: Aug. 27, 2015 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/16 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/02 (2006.01) CI2N 9/78 (2006.01) (71) Applicant: BP Corporation North America Inc., CI2N 9/12 (2006.01) Naperville, IL (US) CI2N 9/24 (2006.01) CI2O 1/02 (2006.01) (72) Inventors: Eric J. Mathur, San Diego, CA (US); CI2N 9/42 (2006.01) Cathy Chang, San Marcos, CA (US) (52) U.S. Cl. CPC. CI2N 9/88 (2013.01); C12O 1/02 (2013.01); (21) Appl. No.: 14/630,006 CI2O I/04 (2013.01): CI2N 9/80 (2013.01); CI2N 9/241.1 (2013.01); C12N 9/0065 (22) Filed: Feb. 24, 2015 (2013.01); C12N 9/2437 (2013.01); C12N 9/14 Related U.S. Application Data (2013.01); C12N 9/16 (2013.01); C12N 9/0061 (2013.01); C12N 9/78 (2013.01); C12N 9/0071 (62) Division of application No. 13/400,365, filed on Feb. (2013.01); C12N 9/1241 (2013.01): CI2N 20, 2012, now Pat. No. 8,962,800, which is a division 9/2482 (2013.01); C07K 2/00 (2013.01); C12Y of application No. 1 1/817,403, filed on May 7, 2008, 305/01004 (2013.01); C12Y 1 1 1/01016 now Pat. No. 8,119,385, filed as application No. PCT/ (2013.01); C12Y302/01004 (2013.01); C12Y US2006/007642 on Mar. 3, 2006. -

A Comprehensive Proteomics and Genomics Analysis Reveals Novel
Supplemental Material can be found at: http://www.mcponline.org/cgi/content/full/D600007-MCP200/ DC1 Dataset A Comprehensive Proteomics and Genomics Analysis Reveals Novel Transmembrane Proteins in Human Platelets and Mouse Megakaryocytes Including G6b-B, a Novel Immunoreceptor Tyrosine-based Inhibitory Motif Protein*□S Yotis A. Senis‡§, Michael G. Tomlinson‡¶, A´ ngel Garcíaʈ**, Stephanie Dumon‡, Victoria L. Heath‡, John Herbert‡, Stephen P. Cobbold‡‡, Jennifer C. Spalton‡, Sinem Ayman§§, Robin Antrobusʈ, Nicole Zitzmannʈ, Roy Bicknell‡, Jon Frampton‡, Downloaded from Kalwant S. Authi§§, Ashley Martin¶¶, Michael J. O. Wakelam¶¶, and Stephen P. Watson‡ʈʈ The platelet surface is poorly characterized due to the low tance of mutant mouse models in establishing protein abundance of many membrane proteins and the lack of function in platelets. This approach identified all of the www.mcponline.org specialist tools for their investigation. In this study we major classes of platelet transmembrane receptors, in- identified novel human platelet and mouse megakaryocyte cluding multitransmembrane proteins. Strikingly 17 of the membrane proteins using specialist proteomics and 25 most megakaryocyte-specific genes (relative to 30 genomics approaches. Three separate methods were used other serial analysis of gene expression libraries) were to enrich platelet surface proteins prior to identification by transmembrane proteins, illustrating the unique nature of liquid chromatography and tandem mass spectrometry: the megakaryocyte/platelet surface. The list of novel by Yotis Senis on April 2, 2007 lectin affinity chromatography, biotin/NeutrAvidin affinity plasma membrane proteins identified using proteomics chromatography, and free flow electrophoresis. Many includes the immunoglobulin superfamily member G6b, known, abundant platelet surface transmembrane pro- which undergoes extensive alternate splicing. -

Mitochondrial Functionality in Male Fertility: from Spermatogenesis to Fertilization
antioxidants Review Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization Yoo-Jin Park and Myung-Geol Pang * Department of Animal Science & Technology and BET Research Institute, Chung-Ang University, Anseong 17546, Gyeonggi-do, Korea; [email protected] * Correspondence: [email protected] Abstract: Mitochondria are structurally and functionally distinct organelles that produce adenosine triphosphate (ATP) through oxidative phosphorylation (OXPHOS), to provide energy to spermatozoa. They can also produce reactive oxidation species (ROS). While a moderate concentration of ROS is critical for tyrosine phosphorylation in cholesterol efflux, sperm–egg interaction, and fertilization, excessive ROS generation is associated with male infertility. Moreover, mitochondria participate in diverse processes ranging from spermatogenesis to fertilization to regulate male fertility. This review aimed to summarize the roles of mitochondria in male fertility depending on the sperm developmen- tal stage (from male reproductive tract to female reproductive tract). Moreover, mitochondria are also involved in testosterone production, regulation of proton secretion into the lumen to maintain an acidic condition in the epididymis, and sperm DNA condensation during epididymal maturation. We also established the new signaling pathway using previous proteomic data associated with male fertility, to understand the overall role of mitochondria in male fertility. The pathway revealed that male infertility is associated with a loss of mitochondrial proteins in spermatozoa, which induces low sperm motility, reduces OXPHOS activity, and results in male infertility. Keywords: mitochondria; oxidative phosphorylation; spermatozoa; male infertility; testis; epididymis; capacitation; fertilization Citation: Park, Y.-J.; Pang, M.-G. Mitochondrial Functionality in Male Fertility: From Spermatogenesis to 1. Introduction Fertilization. Antioxidants 2021, 10, 98. -

Electronic Supplementary Material (ESI) for Molecular Omics. This Journal Is © the Royal Society of Chemistry 2018
Electronic Supplementary Material (ESI) for Molecular Omics. This journal is © The Royal Society of Chemistry 2018 Supplementary Table 1 Biological Rep 1 Biological Rep 2 Biological Rep 3 Fold change w.r.t Control Fold change w.r.t Control Fold change w.r.t Control % CV No. of No. of No. of Acession ID Description peptides Tm 4-PBA 4-PBA+Tm peptides Tm 4-PBA 4-PBA+Tm peptides Tm 4-PBA 4-PBA+Tm Tm 4-PBA 4-PBA+Tm Cytoplasmic dynein 1 heavy chain 1 OS=Homo sapiens GN=DYNC1H1 Q14204 PE=1 SV=5 195 1.04 1.01 1.04 171 0.96 0.98 0.97 165 1.03 1.02 1.03 4.27 2.06 3.81 DNA-dependent protein kinase catalytic subunit OS=Homo sapiens P78527 GN=PRKDC PE=1 SV=3 185 0.93 1.01 0.96 149 0.93 0.99 0.94 139 0.93 0.97 0.95 0.48 2.03 0.92 Q60FE2 MYH9 variant protein OS=Homo sapiens GN=MYH9 PE=2 SV=1 151 1.16 0.99 1.17 138 1.19 1.01 1.20 137 1.15 1.00 1.19 1.49 0.97 1.46 Q60FE6 Filamin A OS=Homo sapiens GN=FLNA PE=2 SV=1 132 1.07 0.98 1.13 119 1.05 0.98 1.14 108 1.07 0.97 1.14 1.01 0.91 0.65 P49327 Fatty acid synthase OS=Homo sapiens GN=FASN PE=1 SV=3 134 0.80 0.96 0.87 96 0.82 0.94 0.87 87 0.83 0.97 0.88 2.09 1.93 0.80 Spectrin beta non-erythrocytic 1 OS=Homo sapiens GN=SPTBN1 PE=2 B2ZZ89 SV=1 103 0.98 0.99 0.97 90 0.94 0.95 0.93 73 1.02 1.04 1.01 4.15 4.78 4.13 Q00610 Clathrin heavy chain 1 OS=Homo sapiens GN=CLTC PE=1 SV=5 144 0.95 1.00 1.02 127 0.94 0.97 0.94 112 0.96 1.02 0.97 0.88 2.61 3.99 Q9Y490 Talin-1 OS=Homo sapiens GN=TLN1 PE=1 SV=3 114 1.03 0.96 1.08 121 0.96 0.91 0.99 81 1.03 1.02 1.03 3.98 5.81 4.13 Peroxisome proliferator activated receptor interacting