Functional Validation of GWAS Gene Candidates for Abnormal Liver Function During Zebrafish Liver Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Genetic Polymorphisms of PNPLA3 and SAMM50 Are Associated with Nonalcoholic Fatty Liver Disease in a Korean Population
Gut and Liver, Vol. 12, No. 3, May 2018, pp. 316-323 ORiginal Article Genetic Polymorphisms of PNPLA3 and SAMM50 Are Associated with Nonalcoholic Fatty Liver Disease in a Korean Population Goh Eun Chung1, Young Lee2, Jeong Yoon Yim1,2, Eun Kyung Choe2, Min-Sun Kwak1, Jong In Yang1, Boram Park3, Jong-Eun Lee4, Jeong A Kim4, and Joo Sung Kim1,5 1Department of Internal Medicine and 2Healthcare Research Institute, Healthcare System Gangnam Center, Seoul National University Hospital, 3Department of Public Health Science, Graduate School of Public Health, Seoul National University, 4DNALink, Inc., and 5Department of Internal Medicine and Liver Research Institute, Seoul National University College of Medicine, Seoul, Korea Background/Aims: The development of nonalcoholic fatty INTRODUCTION liver disease (NAFLD) is associated with multiple genetic and environmental factors. Methods: We performed a genome- Nonalcoholic fatty liver disease (NAFLD) has been recognized wide association study to identify the genetic factors related as the leading cause of chronic liver disease, with a prevalence to NAFLD in a Korean population-based sample of 1,593 up to 20%–30% in the general population.1 Most cases of subjects with NAFLD and 2,816 controls. We replicated the NAFLD follow a benign clinical course, however, once simple data in another sample that included 744 NAFLD patients steatosis progresses to nonalcoholic steatohepatitis (NASH), 25% and 1,137 controls. We investigated single-nucleotide poly- of patients may experience further progression to liver fibrosis morphisms (SNPs) that were related to NAFLD. Results: and cirrhosis.2,3 Furthermore, patients with NASH have increased After adjusting for age, sex and body mass index, rs738409, liver-related mortality compared with the general population.4 rs12483959 and rs2281135, located in the PNPLA3 gene, Genetic and environmental factors have important roles in the were validated in our population (p<8.56×10–8) in the development of NAFLD.5,6 Single-nucleotide polymorphisms same linkage disequilibrium block. -

Proteomic Analysis Reveals a Mitochondrial Remodeling of Βtc3 Cells in Response to Nanotopography
fcell-08-00508 July 29, 2020 Time: 12:23 # 1 ORIGINAL RESEARCH published: 29 July 2020 doi: 10.3389/fcell.2020.00508 Proteomic Analysis Reveals a Mitochondrial Remodeling of bTC3 Cells in Response to Nanotopography Elisa Maffioli1,2†, Alessandra Galli3†, Simona Nonnis1,2, Algerta Marku3, Armando Negri1, Claudio Piazzoni2,4, Paolo Milani2,4, Cristina Lenardi2,4, Carla Perego3* and Gabriella Tedeschi1,2* 1 Department of Veterinary Medicine, University of Milano, Milan, Italy, 2 Centre for Nanostructured Materials and Interfaces, University of Milano, Milan, Italy, 3 Department of Pharmacological and Biomolecular Sciences, University of Milano, Milan, Italy, 4 Department of Physics, University of Milano, Milan, Italy Edited by: Recently, using cluster-assembled zirconia substrates with tailored roughness produced Luisa Pieroni, Santa Lucia Foundation (IRCCS), Italy by supersonic cluster beam deposition, we demonstrated that b cells can sense Reviewed by: nanoscale features of the substrate and can translate these stimuli into a Massimiliano Galluzzi, mechanotransductive pathway capable of preserveing b-cell differentiation and function Chinese Academy of Sciences (CAS), in vitro in long-term cultures of human islets. Using the same proteomic approach, China Geeta Upadhyay, we now focused on the mitochondrial fraction of bTC3 cells grown on the same Uniformed Services University of the zirconia substrates and characterized the morphological and proteomic modifications Health Sciences, United States Gian Maria Fimia, induced by the nanostructure. The results suggest that, in bTC3 cells, mitochondria Sapienza University of Rome, Italy are perturbed by the nanotopography and activate a program involving metabolism *Correspondence: modification and modulation of their interplay with other organelles. Data were confirmed Carla Perego in INS1E, a different b-cell model. -

Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis
Getting “Under the Skin”: Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis by Kristen Monét Brown A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Epidemiological Science) in the University of Michigan 2017 Doctoral Committee: Professor Ana V. Diez-Roux, Co-Chair, Drexel University Professor Sharon R. Kardia, Co-Chair Professor Bhramar Mukherjee Assistant Professor Belinda Needham Assistant Professor Jennifer A. Smith © Kristen Monét Brown, 2017 [email protected] ORCID iD: 0000-0002-9955-0568 Dedication I dedicate this dissertation to my grandmother, Gertrude Delores Hampton. Nanny, no one wanted to see me become “Dr. Brown” more than you. I know that you are standing over the bannister of heaven smiling and beaming with pride. I love you more than my words could ever fully express. ii Acknowledgements First, I give honor to God, who is the head of my life. Truly, without Him, none of this would be possible. Countless times throughout this doctoral journey I have relied my favorite scripture, “And we know that all things work together for good, to them that love God, to them who are called according to His purpose (Romans 8:28).” Secondly, I acknowledge my parents, James and Marilyn Brown. From an early age, you two instilled in me the value of education and have been my biggest cheerleaders throughout my entire life. I thank you for your unconditional love, encouragement, sacrifices, and support. I would not be here today without you. I truly thank God that out of the all of the people in the world that He could have chosen to be my parents, that He chose the two of you. -
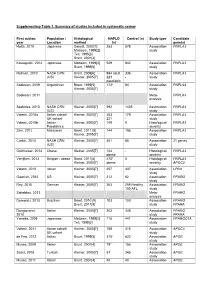
Supplementary Table 1. Summary of Studies Included in Systematic Review
Supplementary Table 1. Summary of studies included in systematic review First author, Population / Histological NAFLD Control (n) Study type Candidate year Location method (n) gene(s) Hotta, 2010 Japanese Sanyal, 2002[1] 253 578 Association PNPLA3 Matteoni, 1999[2] study Teli, 1995[3] Brunt, 2001[4] Kawaguchi, 2012 Japanese Matteoni, 1999[2] 529 942 Association PNPLA3 Brunt, 1999[5] study Rotman, 2010 NASH CRN Brunt, 2009[6] 894 adult 336 Association PNPLA3 (US) Kleiner, 2005[7] 223 - study paediatric Sookoian, 2009 Argentinian Brunt, 1999[5] 172* 94 Association PNPLA3 Kleiner, 2005[7] study Sookoian, 2011 Meta- PNPLA3 analysis Speliotes, 2010 NASH CRN Kleiner, 2005[7] 592 1405 Association PNPLA3 (US) study Valenti, 2010a Italian cohort/ Kleiner, 2005[7] 253 179 Association PNPLA3 UK cohort 321 - study Valenti, 2010b Italian - Kleiner, 2005[7] 149 0 Histological PNPLA3 Paediatrics severity Zain, 2012 Malaysian Brunt, 2011 [8] 144 198 Association PNPLA3 Kleiner, 2005[7] study Corbin, 2013 NASH CRN Kleiner, 2005[7] 361 85 Association 21 genes (US) study Guichelaar, 2013 Obese Kleiner, 2005[7] 144 - Histological PNPLA3 obese severity Verrijken, 2013 Belgian - obese Brunt, 2011[8] 470* 0 Histological PNPLA3 Kleiner, 2005[7] obese severity APOC3 Valenti, 2012 Italian Kleiner, 2005[7] 257 337 Association LPIN1 study Gawrieh, 2012 US Kleiner, 2005[7] 212 62 Association PPARG study Rey, 2010 German Kleiner, 2005[7] 263 259 Healthy Association PPARG 100 AFL study Sahebkar, 2013 Meta- PPARG analysis Domenici, 2013 Brazilian Brunt, 2010 [9] 103 103 -

Targeted Next-Generation Sequencing and Fine Linkage Disequilibrium Mapping Reveals Association of PNPLA3 and PARVB with The
Journal of Human Genetics (2014) 59, 241–246 & 2014 The Japan Society of Human Genetics All rights reserved 1434-5161/14 www.nature.com/jhg ORIGINAL ARTICLE Targeted next-generation sequencing and fine linkage disequilibrium mapping reveals association of PNPLA3 and PARVB with the severity of nonalcoholic fatty liver disease Takuya Kitamoto1, Aya Kitamoto1, Masato Yoneda2, Hideyuki Hyogo3, Hidenori Ochi3, Seiho Mizusawa1, Takato Ueno4, Kazuwa Nakao5, Akihiro Sekine1, Kazuaki Chayama3, Atsushi Nakajima2 and Kikuko Hotta1 The genomic regions containing PNPLA3, SAMM50 and PARVB are susceptibility loci for the development and progression of nonalcoholic fatty liver disease (NAFLD). In order to search for all common variations in this region, we amplified the genomic DNA of 28 NAFLD patients by long-range PCR, covering the entire susceptibility region and sequenced the DNA using indexed multiplex next-generation sequencing. We found 329 variations, including four novel variations. Fine mapping of variations including insertion/deletions was performed for 540 NAFLD patients (488 with nonalcoholic steatohepatitis (NASH) and 52 with simple steatosis) and 1012 control subjects. HaploView analysis showed that linkage disequilibrium (LD) block 1 and 2 occurred in PNPLA3, block 3 in SAMM50 and block 4 in PARVB. Variations in LD blocks 1–4 were significantly associated with NAFLD as compared with control subjects (Po1 Â 10 À8). Variations in LD block 2 were significantly associated with the NAFLD activity score (NAS), aspartate aminotransferase and alanine aminotransferase. Variations in LD block 1 were significantly associated with the fibrosis stage. The strongest associations were observed for variations in LD block 4, with NASH as compared with simple steatosis (P ¼ 7.1 Â 10 À6) and NAS (P ¼ 3.4 Â 10 À6). -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

UNIVERSITY of CALIFORNIA, SAN DIEGO Measuring
UNIVERSITY OF CALIFORNIA, SAN DIEGO Measuring and Correlating Blood and Brain Gene Expression Levels: Assays, Inbred Mouse Strain Comparisons, and Applications to Human Disease Assessment A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Biomedical Sciences by Mary Elizabeth Winn Committee in charge: Professor Nicholas J Schork, Chair Professor Gene Yeo, Co-Chair Professor Eric Courchesne Professor Ron Kuczenski Professor Sanford Shattil 2011 Copyright Mary Elizabeth Winn, 2011 All rights reserved. 2 The dissertation of Mary Elizabeth Winn is approved, and it is acceptable in quality and form for publication on microfilm and electronically: Co-Chair Chair University of California, San Diego 2011 iii DEDICATION To my parents, Dennis E. Winn II and Ann M. Winn, to my siblings, Jessica A. Winn and Stephen J. Winn, and to all who have supported me throughout this journey. iv TABLE OF CONTENTS Signature Page iii Dedication iv Table of Contents v List of Figures viii List of Tables x Acknowledgements xiii Vita xvi Abstract of Dissertation xix Chapter 1 Introduction and Background 1 INTRODUCTION 2 Translational Genomics, Genome-wide Expression Analysis, and Biomarker Discovery 2 Neuropsychiatric Diseases, Tissue Accessibility and Blood-based Gene Expression 4 Mouse Models of Human Disease 5 Microarray Gene Expression Profiling and Globin Reduction 7 Finding and Accessible Surrogate Tissue for Neural Tissue 9 Genetic Background Effect Analysis 11 SPECIFIC AIMS 12 ENUMERATION OF CHAPTERS -

A Network Inference Approach to Understanding Musculoskeletal
A NETWORK INFERENCE APPROACH TO UNDERSTANDING MUSCULOSKELETAL DISORDERS by NIL TURAN A thesis submitted to The University of Birmingham for the degree of Doctor of Philosophy College of Life and Environmental Sciences School of Biosciences The University of Birmingham June 2013 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Musculoskeletal disorders are among the most important health problem affecting the quality of life and contributing to a high burden on healthcare systems worldwide. Understanding the molecular mechanisms underlying these disorders is crucial for the development of efficient treatments. In this thesis, musculoskeletal disorders including muscle wasting, bone loss and cartilage deformation have been studied using systems biology approaches. Muscle wasting occurring as a systemic effect in COPD patients has been investigated with an integrative network inference approach. This work has lead to a model describing the relationship between muscle molecular and physiological response to training and systemic inflammatory mediators. This model has shown for the first time that oxygen dependent changes in the expression of epigenetic modifiers and not chronic inflammation may be causally linked to muscle dysfunction. -

1 a Framework to Identify Modifier Genes in Patients With
bioRxiv preprint doi: https://doi.org/10.1101/117978; this version posted March 18, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. A framework to identify modifier genes in patients with Phelan-McDermid syndrome Short running title: Mapping modifier genes in Phelan-McDermid Syndrome Anne-Claude Tabet1,2,3,4¶, Thomas Rolland2,3,4¶, Marie Ducloy2,3,4, Jonathan Lévy1, Julien Buratti2,3,4, Alexandre Mathieu2,3,4, Damien Haye1, Laurence Perrin1, Céline Dupont1, Sandrine Passemard1, Yline Capri1, Alain Verloes1, Séverine Drunat1, Boris Keren5, Cyril Mignot6, Isabelle Marey7, Aurélia Jacquette7, Sandra Whalen7, Eva Pipiras8, Brigitte Benzacken8, Sandra Chantot-Bastaraud9, Alexandra Afenjar10, Delphine Héron10, Cédric Le Caignec11, Claire Beneteau11, Olivier Pichon11, Bertrand Isidor11, Albert David11, Jean-Michel Dupont12, Stephan Kemeny13, Laetitia Gouas13, Philippe Vago13, Anne-Laure Mosca-Boidron14, Laurence Faivre15, Chantal Missirian16, Nicole Philip16, Damien Sanlaville17, Patrick Edery18, Véronique Satre19, Charles Coutton19, Françoise Devillard19, Klaus Dieterich20, Marie-Laure Vuillaume21, Caroline Rooryck21, Didier Lacombe21, Lucile Pinson22, Vincent Gatinois22, Jacques Puechberty22, Jean Chiesa23, James Lespinasse24, Christèle Dubourg25, Chloé Quelin25, Mélanie Fradin25, Hubert Journel26, Annick Toutain27, Dominique Martin28, Abdelamdjid Benmansour1, -

Identification of Differentially Methylated Region (DMR) Networks Associated with Progression of Nonalcoholic Fatty Liver Disease
www.nature.com/scientificreports OPEN Identifcation of diferentially methylated region (DMR) networks associated with progression of Received: 16 March 2018 Accepted: 29 August 2018 nonalcoholic fatty liver disease Published: xx xx xxxx Kikuko Hotta 1, Aya Kitamoto2, Takuya Kitamoto2, Yuji Ogawa3, Yasushi Honda3, Takaomi Kessoku3, Masato Yoneda3, Kento Imajo3, Wataru Tomeno3,4, Satoru Saito3 & Atsushi Nakajima3 The progression of nonalcoholic fatty liver disease (NAFLD) is afected by epigenetics. We performed diferentially methylated region (DMR) and co-methylation analyses to identify DMR networks associated with the progression of NAFLD. DMRs displaying diferences in multiple consecutive diferentially methylated CpGs between mild and advanced NAFLD were extracted. The average values of topological overlap measures for the CpG matrix combining two diferent DMRs were calculated and two DMR networks that strongly correlated with the stages of fbrosis were identifed. The annotated genes of one network included genes involved in transcriptional regulation, cytoskeleton organization, and cellular proliferation. The annotated genes of the second network were primarily associated with metabolic pathways. The CpG methylation levels in these networks were strongly afected by age and fasting plasma glucose levels, which may be important co-regulatory factors. The methylation status of fve DMRs in the second network was reversible following weight loss. Our results suggest that CpG methylation in DMR networks is regulated concomitantly via aging