Proteomic Analysis Reveals a Mitochondrial Remodeling of Βtc3 Cells in Response to Nanotopography
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Validation of GWAS Gene Candidates for Abnormal Liver Function During Zebrafish Liver Development
Disease Models & Mechanisms 6, 1271-1278 (2013) doi:10.1242/dmm.011726 RESEARCH REPORT Functional validation of GWAS gene candidates for abnormal liver function during zebrafish liver development Leah Y. Liu1, Caroline S. Fox2,3, Trista E. North4,5 and Wolfram Goessling1,5,6,7,* SUMMARY Genome-wide association studies (GWAS) have revealed numerous associations between many phenotypes and gene candidates. Frequently, however, further elucidation of gene function has not been achieved. A recent GWAS identified 69 candidate genes associated with elevated liver enzyme concentrations, which are clinical markers of liver disease. To investigate the role of these genes in liver homeostasis, we narrowed down this list to 12 genes based on zebrafish orthology, zebrafish liver expression and disease correlation. To assess the function of gene candidates during liver development, we assayed hepatic progenitors at 48 hours post fertilization (hpf) and hepatocytes at 72 hpf using in situ hybridization following morpholino knockdown in zebrafish embryos. Knockdown of three genes (pnpla3, pklr and mapk10) decreased expression of hepatic progenitor cells, whereas knockdown of eight genes (pnpla3, cpn1, trib1, fads2, slc2a2, pklr, mapk10 and samm50) decreased cell-specific hepatocyte expression. We then induced liver injury in zebrafish embryos using acetaminophen exposure and observed changes in liver toxicity incidence in morphants. Prioritization of GWAS candidates and morpholino knockdown expedites the study of newly identified genes impacting liver development and represents a feasible method for initial assessment of candidate genes to instruct further mechanistic analyses. Our analysis can be extended to DMM GWAS for additional disease-associated phenotypes. INTRODUCTION of 61,089 individuals, which increased the likelihood that additional Levels of liver enzymes such as alanine aminotransferase (ALT), genes reached statistical significance (Chambers et al., 2011). -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -

Comprehensive Identification and Characterization of Somatic Copy Number Alterations in Triple‑Negative Breast Cancer
INTERNATIONAL JOURNAL OF ONCOLOGY 56: 522-530, 2020 Comprehensive identification and characterization of somatic copy number alterations in triple‑negative breast cancer ZAIBING LI1,2*, XIAO ZHANG3*, CHENXIN HOU4, YUQING ZHOU4, JUNLI CHEN1, HAOYANG CAI5, YIFENG YE3, JINPING LIU3 and NING HUANG1 1Department of Pathophysiology, West China School of Basic Medical Sciences and Forensic Medicine, Sichuan University, Chengdu, Sichuan 610041; 2Department of Pathophysiology, School of Basic Medical Science, Southwest Medical University, Luzhou, Sichuan 646000; 3Department of Breast Surgery, Sichuan Provincial People's Hospital, University of Electronic Science and Technology of China, Chengdu, Sichuan 611731; 4West China Medical School, Sichuan University, Chengdu, Sichuan 610041; 5Center of Growth, Metabolism and Aging, Key Laboratory of Bio‑Resources and Eco‑Environment, College of Life Sciences, Sichuan University, Chengdu, Sichuan 610064, P.R. China Received January 30, 2019; Accepted August 30, 2019 DOI: 10.3892/ijo.2019.4950 Abstract. Triple-negative breast cancer (TNBC) accounts hierarchical clustering of tumors resulted in three main for ~15% of all breast cancer diagnoses each year. Patients subgroups that exhibited distinct CNA profiles, which with TNBC tend to have a higher risk for early relapse and may reveal the heterogeneity of molecular mechanisms in a worse prognosis. TNBC is characterized by extensive TNBC subgroups. These results will extend the molecular somatic copy number alterations (CNAs). However, the DNA understanding of TNBC and will facilitate the discovery of CNA profile of TNBC remains to be extensively investigated. therapeutic and diagnostic target candidates. The present study assessed the genomic profile of CNAs in 201 TNBC samples, aiming to identify recurrent CNAs that Introduction may drive the pathogenesis of TNBC. -

Coordinate Regulation of Long Non-Coding Rnas and Protein-Coding Genes in Germ- Free Mice Joseph Dempsey, Angela Zhang and Julia Yue Cui*
Dempsey et al. BMC Genomics (2018) 19:834 https://doi.org/10.1186/s12864-018-5235-3 RESEARCHARTICLE Open Access Coordinate regulation of long non-coding RNAs and protein-coding genes in germ- free mice Joseph Dempsey, Angela Zhang and Julia Yue Cui* Abstract Background: Long non-coding RNAs (lncRNAs) are increasingly recognized as regulators of tissue-specific cellular functions and have been shown to regulate transcriptional and translational processes, acting as signals, decoys, guides, and scaffolds. It has been suggested that some lncRNAs act in cis to regulate the expression of neighboring protein-coding genes (PCGs) in a mechanism that fine-tunes gene expression. Gut microbiome is increasingly recognized as a regulator of development, inflammation, host metabolic processes, and xenobiotic metabolism. However, there is little known regarding whether the gut microbiome modulates lncRNA gene expression in various host metabolic organs. The goals of this study were to 1) characterize the tissue-specific expression of lncRNAs and 2) identify and annotate lncRNAs differentially regulated in the absence of gut microbiome. Results: Total RNA was isolated from various tissues (liver, duodenum, jejunum, ileum, colon, brown adipose tissue, white adipose tissue, and skeletal muscle) from adult male conventional and germ-free mice (n = 3 per group). RNA-Seq was conducted and reads were mapped to the mouse reference genome (mm10) using HISAT. Transcript abundance and differential expression was determined with Cufflinks using the reference databases NONCODE 2016 for lncRNAs and UCSC mm10 for PCGs. Although the constitutive expression of lncRNAs was ubiquitous within the enterohepatic (liver and intestine) and the peripheral metabolic tissues (fat and muscle) in conventional mice, differential expression of lncRNAs by lack of gut microbiota was highly tissue specific. -

Antisense Afp Transcripts in Mouse Liver and Their Potential Role in Afp Gene Regulation
University of Kentucky UKnowledge Theses and Dissertations--Microbiology, Microbiology, Immunology, and Molecular Immunology, and Molecular Genetics Genetics 2017 ANTISENSE AFP TRANSCRIPTS IN MOUSE LIVER AND THEIR POTENTIAL ROLE IN AFP GENE REGULATION Maria S. Dixon University of Kentucky, [email protected] Digital Object Identifier: https://doi.org/10.13023/ETD.2017.356 Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Dixon, Maria S., "ANTISENSE AFP TRANSCRIPTS IN MOUSE LIVER AND THEIR POTENTIAL ROLE IN AFP GENE REGULATION" (2017). Theses and Dissertations--Microbiology, Immunology, and Molecular Genetics. 14. https://uknowledge.uky.edu/microbio_etds/14 This Doctoral Dissertation is brought to you for free and open access by the Microbiology, Immunology, and Molecular Genetics at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Microbiology, Immunology, and Molecular Genetics by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Genetic Polymorphisms of PNPLA3 and SAMM50 Are Associated with Nonalcoholic Fatty Liver Disease in a Korean Population
Gut and Liver, Vol. 12, No. 3, May 2018, pp. 316-323 ORiginal Article Genetic Polymorphisms of PNPLA3 and SAMM50 Are Associated with Nonalcoholic Fatty Liver Disease in a Korean Population Goh Eun Chung1, Young Lee2, Jeong Yoon Yim1,2, Eun Kyung Choe2, Min-Sun Kwak1, Jong In Yang1, Boram Park3, Jong-Eun Lee4, Jeong A Kim4, and Joo Sung Kim1,5 1Department of Internal Medicine and 2Healthcare Research Institute, Healthcare System Gangnam Center, Seoul National University Hospital, 3Department of Public Health Science, Graduate School of Public Health, Seoul National University, 4DNALink, Inc., and 5Department of Internal Medicine and Liver Research Institute, Seoul National University College of Medicine, Seoul, Korea Background/Aims: The development of nonalcoholic fatty INTRODUCTION liver disease (NAFLD) is associated with multiple genetic and environmental factors. Methods: We performed a genome- Nonalcoholic fatty liver disease (NAFLD) has been recognized wide association study to identify the genetic factors related as the leading cause of chronic liver disease, with a prevalence to NAFLD in a Korean population-based sample of 1,593 up to 20%–30% in the general population.1 Most cases of subjects with NAFLD and 2,816 controls. We replicated the NAFLD follow a benign clinical course, however, once simple data in another sample that included 744 NAFLD patients steatosis progresses to nonalcoholic steatohepatitis (NASH), 25% and 1,137 controls. We investigated single-nucleotide poly- of patients may experience further progression to liver fibrosis morphisms (SNPs) that were related to NAFLD. Results: and cirrhosis.2,3 Furthermore, patients with NASH have increased After adjusting for age, sex and body mass index, rs738409, liver-related mortality compared with the general population.4 rs12483959 and rs2281135, located in the PNPLA3 gene, Genetic and environmental factors have important roles in the were validated in our population (p<8.56×10–8) in the development of NAFLD.5,6 Single-nucleotide polymorphisms same linkage disequilibrium block. -

Epigenetic Regulation of WNT3A Enhancer During Regeneration of Injured Cortical Neurons
International Journal of Molecular Sciences Article Epigenetic Regulation of WNT3A Enhancer during Regeneration of Injured Cortical Neurons 1, 2, 3 2 1 Chu-Yuan Chang y, Jui-Hung Hung y, Liang-Wei Huang , Joye Li , Ka Shing Fung , Cheng-Fu Kao 4 and Linyi Chen 1,5,* 1 Institute of Molecular Medicine, National Tsing Hua University, Hsinchu 30013, Taiwan; [email protected] (C.-Y.C.); [email protected] (K.S.F.) 2 Department of Computer Science, National Chiao Tung University, Hsinchu 30010, Taiwan; [email protected] (J.-H.H.); [email protected] (J.L.) 3 Department of Life Science, National Tsing Hua University, Hsinchu 30013, Taiwan; [email protected] 4 Institute of Cellular and Organismic Biology, Academia Sinica, Taipei 11574, Taiwan; [email protected] 5 Department of Medical Science, National Tsing Hua University, Hsinchu 30013, Taiwan * Correspondence: [email protected]; Tel.: +886-3-574-2775; Fax: +886-3-571-5934 These authors contributed equally to this work. y Received: 21 February 2020; Accepted: 9 March 2020; Published: 10 March 2020 Abstract: Traumatic brain injury is known to reprogram the epigenome. Chromatin immunoprecipitation-sequencing of histone H3 lysine 27 acetylation (H3K27ac) and tri-methylation of histone H3 at lysine 4 (H3K4me3) marks was performed to address the transcriptional regulation of candidate regeneration-associated genes. In this study, we identify a novel enhancer region for induced WNT3A transcription during regeneration of injured cortical neurons. We further demonstrated an increased mono-methylation of histone H3 at lysine 4 (H3K4me1) modification at this enhancer concomitant with a topological interaction between sub-regions of this enhancer and with promoter of WNT3A gene. -

Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis
Getting “Under the Skin”: Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis by Kristen Monét Brown A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Epidemiological Science) in the University of Michigan 2017 Doctoral Committee: Professor Ana V. Diez-Roux, Co-Chair, Drexel University Professor Sharon R. Kardia, Co-Chair Professor Bhramar Mukherjee Assistant Professor Belinda Needham Assistant Professor Jennifer A. Smith © Kristen Monét Brown, 2017 [email protected] ORCID iD: 0000-0002-9955-0568 Dedication I dedicate this dissertation to my grandmother, Gertrude Delores Hampton. Nanny, no one wanted to see me become “Dr. Brown” more than you. I know that you are standing over the bannister of heaven smiling and beaming with pride. I love you more than my words could ever fully express. ii Acknowledgements First, I give honor to God, who is the head of my life. Truly, without Him, none of this would be possible. Countless times throughout this doctoral journey I have relied my favorite scripture, “And we know that all things work together for good, to them that love God, to them who are called according to His purpose (Romans 8:28).” Secondly, I acknowledge my parents, James and Marilyn Brown. From an early age, you two instilled in me the value of education and have been my biggest cheerleaders throughout my entire life. I thank you for your unconditional love, encouragement, sacrifices, and support. I would not be here today without you. I truly thank God that out of the all of the people in the world that He could have chosen to be my parents, that He chose the two of you. -
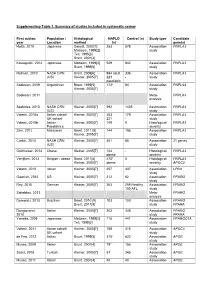
Supplementary Table 1. Summary of Studies Included in Systematic Review
Supplementary Table 1. Summary of studies included in systematic review First author, Population / Histological NAFLD Control (n) Study type Candidate year Location method (n) gene(s) Hotta, 2010 Japanese Sanyal, 2002[1] 253 578 Association PNPLA3 Matteoni, 1999[2] study Teli, 1995[3] Brunt, 2001[4] Kawaguchi, 2012 Japanese Matteoni, 1999[2] 529 942 Association PNPLA3 Brunt, 1999[5] study Rotman, 2010 NASH CRN Brunt, 2009[6] 894 adult 336 Association PNPLA3 (US) Kleiner, 2005[7] 223 - study paediatric Sookoian, 2009 Argentinian Brunt, 1999[5] 172* 94 Association PNPLA3 Kleiner, 2005[7] study Sookoian, 2011 Meta- PNPLA3 analysis Speliotes, 2010 NASH CRN Kleiner, 2005[7] 592 1405 Association PNPLA3 (US) study Valenti, 2010a Italian cohort/ Kleiner, 2005[7] 253 179 Association PNPLA3 UK cohort 321 - study Valenti, 2010b Italian - Kleiner, 2005[7] 149 0 Histological PNPLA3 Paediatrics severity Zain, 2012 Malaysian Brunt, 2011 [8] 144 198 Association PNPLA3 Kleiner, 2005[7] study Corbin, 2013 NASH CRN Kleiner, 2005[7] 361 85 Association 21 genes (US) study Guichelaar, 2013 Obese Kleiner, 2005[7] 144 - Histological PNPLA3 obese severity Verrijken, 2013 Belgian - obese Brunt, 2011[8] 470* 0 Histological PNPLA3 Kleiner, 2005[7] obese severity APOC3 Valenti, 2012 Italian Kleiner, 2005[7] 257 337 Association LPIN1 study Gawrieh, 2012 US Kleiner, 2005[7] 212 62 Association PPARG study Rey, 2010 German Kleiner, 2005[7] 263 259 Healthy Association PPARG 100 AFL study Sahebkar, 2013 Meta- PPARG analysis Domenici, 2013 Brazilian Brunt, 2010 [9] 103 103 -

Examination of Testicular Gene Expression Patterns in Yorkshire Pigs with High and Low Levels of Boar Taint
Animal Biotechnology, 21: 77–87, 2010 Copyright # Taylor & Francis Group, LLC ISSN: 1049-5398 print=1532-2378 online DOI: 10.1080/10495390903500607 EXAMINATION OF TESTICULAR GENE EXPRESSION PATTERNS IN YORKSHIRE PIGS WITH HIGH AND LOW LEVELS OF BOAR TAINT Maxwell C. K. Leung, Kiera-Lynne Bowley, and E. James Squires Department of Animal and Poultry Science, University of Guelph, Guelph, Ontario, Canada Boar taint refers to the objectionable odor and flavor in meat of some uncastrated male pigs, which is primarily due to high levels of androstenone, a steroid produced in the testis, and 3-methylindole (skatole) which is produced by bacterial degradation of tryptophan in the intestinal tract. We determined testicular gene expression patterns of Yorkshire pigs with high and low levels of boar taint using swine DNA microarrays with two-color hybridization. The microarrays contained 19486 annotated probes; the expressions of 8719 genes were detected. Fifty-three genes were significantly up-regulated in the high boar taint group and four were significantly down-regulated (p < 0.05; fold change > Æ1.55). Gene ontology (GO) analysis short-listed 11 significant GO terms (p < 0.05), most of which are associated with steroid metabolism and mitochondrial components. Comparing the results of this study with published work on Duroc and Norwegian Landrace boars,1 eleven genes (HSB17B4, FDX1, CYP11A1, DHRS4, PRDX1, CYB5, CYP17A1, FTL, IDI1, SULT2A1, and RDH12) were over-expressed in all three breeds with a high androstenone level. The current findings confirmed a number of candidate genes identified in previous functional studies and suggest several new genes differentially expressed with different levels of boar taint. -

Targeted Next-Generation Sequencing and Fine Linkage Disequilibrium Mapping Reveals Association of PNPLA3 and PARVB with The
Journal of Human Genetics (2014) 59, 241–246 & 2014 The Japan Society of Human Genetics All rights reserved 1434-5161/14 www.nature.com/jhg ORIGINAL ARTICLE Targeted next-generation sequencing and fine linkage disequilibrium mapping reveals association of PNPLA3 and PARVB with the severity of nonalcoholic fatty liver disease Takuya Kitamoto1, Aya Kitamoto1, Masato Yoneda2, Hideyuki Hyogo3, Hidenori Ochi3, Seiho Mizusawa1, Takato Ueno4, Kazuwa Nakao5, Akihiro Sekine1, Kazuaki Chayama3, Atsushi Nakajima2 and Kikuko Hotta1 The genomic regions containing PNPLA3, SAMM50 and PARVB are susceptibility loci for the development and progression of nonalcoholic fatty liver disease (NAFLD). In order to search for all common variations in this region, we amplified the genomic DNA of 28 NAFLD patients by long-range PCR, covering the entire susceptibility region and sequenced the DNA using indexed multiplex next-generation sequencing. We found 329 variations, including four novel variations. Fine mapping of variations including insertion/deletions was performed for 540 NAFLD patients (488 with nonalcoholic steatohepatitis (NASH) and 52 with simple steatosis) and 1012 control subjects. HaploView analysis showed that linkage disequilibrium (LD) block 1 and 2 occurred in PNPLA3, block 3 in SAMM50 and block 4 in PARVB. Variations in LD blocks 1–4 were significantly associated with NAFLD as compared with control subjects (Po1 Â 10 À8). Variations in LD block 2 were significantly associated with the NAFLD activity score (NAS), aspartate aminotransferase and alanine aminotransferase. Variations in LD block 1 were significantly associated with the fibrosis stage. The strongest associations were observed for variations in LD block 4, with NASH as compared with simple steatosis (P ¼ 7.1 Â 10 À6) and NAS (P ¼ 3.4 Â 10 À6).