Computational Learning of Stem Cell Fates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

High-Fat Diet-Induced Complement Activation Mediates Intestinal Inflammation and Neoplasia, Independent of Obesity Stephanie K
Published OnlineFirst August 17, 2016; DOI: 10.1158/1541-7786.MCR-16-0153 Metabolism Molecular Cancer Research High-Fat Diet-Induced Complement Activation Mediates Intestinal Inflammation and Neoplasia, Independent of Obesity Stephanie K. Doerner1, Edimara S. Reis2, Elaine S. Leung3, Justine S. Ko1, Jason D. Heaney4,5, Nathan A. Berger1,6, John D. Lambris2, and Joseph H. Nadeau1,3 Abstract Obesity and related metabolic disturbances are closely asso- and generation of C5a, which in turn induces the production of ciated with pathologies that represent a significant burden to proinflammatory cytokines and expression of proto-oncogenes. global health. Epidemiological and molecular evidence links Pharmacological and genetic targeting of the C5a receptor obesity and metabolic status with inflammation and increased reduced both inflammation and intestinal polyposis, suggest- risk of cancer. Here, using a mouse model of intestinal neo- ing the use of complement inhibitors for preventing diet- plasia and strains that are susceptible or resistant to diet- induced neoplasia. induced obesity, it is demonstrated that high-fat diet-induced inflammation, rather than obesity or metabolic status, is asso- Implications: This study characterizes the relations between diet ciated with increased intestinal neoplasia. The complement and metabolic conditions on risk for a common cancer and fragmentC5aactsasthetriggerforinflammation and intestinal identifies complement activation as a novel target for cancer tumorigenesis. High-fat diet induces complement activation prevention. Mol Cancer Res; 14(10); 953–65. Ó2016 AACR. Introduction Supporting a role for diet-induced obesity (DIO) in promoting inflammation and cancer, levels of IL6 and tumor burden are both Obesity is an increasingly important risk factor for many ob ob reduced following azoxymethane treatment of obese Lep / mice cancers (1, 2). -

Leptin As a Uremic Toxin Interferes with Neutrophil Chemotaxis
J Am Soc Nephrol 15: 2366–2372, 2004 Leptin as a Uremic Toxin Interferes with Neutrophil Chemotaxis LUCIANO OTTONELLO,* PAOLA GNERRE,* MARIA BERTOLOTTO,* MARINA MANCINI,* PATRIZIA DAPINO,* RODOLFO RUSSO,† GIACOMO GARIBOTTO,† TOMMASO BARRECA,* and FRANCO DALLEGRI* *Division of Internal Medicine and †Division of Nephrology, Department of Internal Medicine and Medical Specialties, University of Genoa Medical School, Genoa, Italy Abstract. Leptin is a pleiotropic molecule involved in energy Finally, the serum inhibitory activity can be effectively prevented homeostasis, hematopoiesis, inflammation, and immunity. Hypo- by immune depletion of leptin. The results also show, however, leptinemia characterizing starvation has been strictly related to that leptin by itself is endowed with chemotactic activity toward increased susceptibility to infection secondary to malnutrition. neutrophils. The two activities—inhibition of the cell response to Nevertheless, ESRD is characterized by high susceptibility to chemokines and stimulation of neutrophil migration—could be bacterial infection despite hyperleptinemia. Defects in neutrophils detected at similar concentrations. On the contrary, neutrophils 2ϩ play a crucial role in the infectious morbidity, and several uremic exposed to leptin did not display detectable [Ca ]i mobilization,  toxins that are capable of depressing neutrophil functions have oxidant production, or 2-integrin upregulation. The results dem- been identified. Only a few and contrasting reports about leptin onstrate that leptin is -
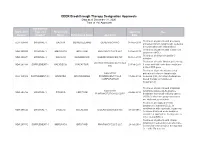
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Obesity and Sex Affect the Immune Responses to Tick-Borne Encephalitis Booster Vaccination
ORIGINAL RESEARCH published: 27 May 2020 doi: 10.3389/fimmu.2020.00860 Obesity and Sex Affect the Immune Responses to Tick-Borne Encephalitis Booster Vaccination Erika Garner-Spitzer 1*†, Eva-Maria Poellabauer 1†, Angelika Wagner 1, Angela Guzek 1, Ines Zwazl 1, Claudia Seidl-Friedrich 1, Christoph J. Binder 2, Karin Stiasny 3, Michael Kundi 4 and Ursula Wiedermann 1* 1 Institute of Specific Prophylaxis and Tropical Medicine, Medical University of Vienna, Vienna, Austria, 2 Department for Laboratory Medicine, Medical University Vienna, Vienna, Austria, 3 Center of Virology, Medical University Vienna, Vienna, Edited by: Austria, 4 Center for Public Health, Medical University Vienna, Vienna, Austria Karl Ljungberg, Eurocine Vaccines AB, Sweden Reviewed by: Obesity has dramatically increased over the last 30 years and reaches according to World Liubov I. Kozlovskaya, Health Organization dimensions of a global epidemic. The obesity-associated chronic Chumakov Federal Scientific Center for Research and Development of low-level inflammation contributes to severe comorbidities and directly affects many Immune-and- Biological Products immune cells leading to immune dysfunction and increased susceptibility to infections. (RAS), Russia Thus, prophylaxis against vaccine-preventable diseases is crucial, yet the responsiveness Galina Grigirievna Karganova, Chumakov Institute of Poliomyelitis to several vaccines is unclear under obesity. In order to assess the responsiveness and Viral Encephalitides (RAS), Russia to tick-borne encephalitis (TBE) vaccine, we revaccinated 37 obese individuals and *Correspondence: 36 normal-weight controls with a licensed TBE vaccine. Metabolic, hormonal, and Erika Garner-Spitzer [email protected] immunologic profiles along with vaccine-specific humoral and cellular immune responses Ursula Wiedermann were evaluated in sera and peripheral blood mononuclear cells (PBMCs) before, 1 week, [email protected] 4 weeks, and 6 months after TBE booster. -

Insulin and Leptin As Adiposity Signals
Insulin and Leptin as Adiposity Signals STEPHEN C. BENOIT,DEBORAH J. CLEGG,RANDY J. SEELEY, AND STEPHEN C. WOODS Department of Psychiatry, University of Cincinnati Medical Center, Cincinnati, Ohio 45267 ABSTRACT There is now considerable consensus that the adipocyte hormone leptin and the pancreatic hormone insulin are important regulators of food intake and energy balance. Leptin and insulin fulfill many of the requirements to be putative adiposity signals to the brain. Plasma leptin and insulin levels are positively correlated with body weight and with adipose mass in particular. Furthermore, both leptin and insulin enter the brain from the plasma. The brain expresses both insulin and leptin receptors in areas important in the control of food intake and energy balance. Consistent with their roles as adiposity signals, exogenous leptin and insulin both reduce food intake when administered locally into the brain in a number of species under different experimental paradigms. Additionally, central administration of insulin antibodies increases food intake and body weight. Recent studies have demonstrated that both insulin and leptin have additive effects when administered simulta- neously. Finally, we recently have demonstrated that leptin and insulin share downstream neuropep- tide signaling pathways. Hence, insulin and leptin provide important negative feedback signals to the central nervous system, proportional to peripheral energy stores and coupled with catabolic circuits. I. Overview When maintained on an ad libitum diet, most animals — including humans — are able to precisely match caloric intake with caloric expenditure, resulting in relatively stable energy stores as adipose tissue (Kennedy, 1953; Keesey, 1986). Growing emphasis has been placed on the role of the central nervous system (CNS) in controlling this precision of energy homeostasis. -

Relevance of Serum Leptin and Leptin-Receptor Concentrations in Critically Ill Patients
Hindawi Publishing Corporation Mediators of Inflammation Volume 2010, Article ID 473540, 9 pages doi:10.1155/2010/473540 Clinical Study Relevance of Serum Leptin and Leptin-Receptor Concentrations in Critically Ill Patients Alexander Koch,1 Ralf Weiskirchen,2 Henning W. Zimmermann,1 Edouard Sanson,1 Christian Trautwein,1 and Frank Tacke1 1 Department of Medicine III, RWTH-University Hospital Aachen, Pauwelsstraße 30, 52074 Aachen, Germany 2 Institute of Clinical Chemistry and Pathobiochemistry, RWTH-University Hospital Aachen, Pauwelsstraße 30, 52074 Aachen, Germany Correspondence should be addressed to Alexander Koch, [email protected] Received 10 March 2010; Accepted 4 May 2010 Academic Editor: Oreste Gualillo Copyright © 2010 Alexander Koch et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The adipocyte-derived cytokine leptin was implicated to link inflammation and metabolic alterations. We investigated the potential role of leptin components in critically ill patients, because systemic inflammation, insulin resistance, and hyperglycemia are common features of critical illness. Upon admission to Medical Intensive Care Unit (ICU), free leptin and soluble leptin-receptor serum concentrations were determined in 137 critically ill patients (95 with sepsis, 42 without sepsis) and 26 healthy controls. Serum leptin or leptin-receptor did not differ between patients or controls and were independent of sepsis. However, serum leptin was closely associated with obesity and diabetes and clearly correlated with markers of metabolism and liver function. Leptin- receptor was an unfavourable prognostic indicator, associated with mortality during three years follow-up. -

Leptin Promotes Glioblastoma Johnathan E. Lawrence1,2, Nicholas J. Cook1,2, Richard A. Rovin2,3, Robert J. Winn1,2 1Biology Depa
Leptin Promotes Glioblastoma Johnathan E. Lawrence1,2, Nicholas J. Cook1,2, Richard A. Rovin2,3, Robert J. Winn1,2 1Biology Department, Northern Michigan University, Marquette, MI 2Upper Michigan Brain Tumor Center, Marquette, MI 3Neurosurgery, Marquette General Hospital, Marquette, MI Address for Correspondence: Robert Winn, Ph.D. Biology Department Northern Michigan University 1401Presque Isle Ave Marquette, MI 49855 Phone: (906) 227-2310 Fax: (906) 227-1063 Email: [email protected] ABSTRACT The hormone leptin has a variety of functions. Originally known for its role in satiety and weight loss, leptin more recently has been shown to augment tumor growth in a variety of cancers. Within gliomas, there is a correlation between tumor grade and tumor expression of leptin and its receptor. This suggests that autocrine signaling within the tumor microenvironment may promote the growth of high grade gliomas. Leptin does this through stimulation of cellular pathways that are also advantageous for tumor growth and recurrence: antiapoptosis, proliferation, angiogenesis and migration. Conversely, a loss of leptin expression attenuates tumor growth. In animal models of colon cancer and melanoma, a decline in the expression and secretion of leptin resulted in a reduction of tumor growth. In these models, positive mental stimulation through environmental enrichment decreased leptin secretion and improved tumor outcome. This review explores the link between leptin and glioblastoma. INTRODUCTION Leptin is the product of the obese gene, located on chromosome 7 in humans. Mice with mutation in the obese gene are obese and insatiable [1]. When exogenous leptin is injected into leptin deficient obese mice (ob/ob mice), the protein promotes satiety and weight loss [2-5]. -

Leptin Induces Mitosis and Activates the Canonical Wnt/ß-Catenin
ORIGINAL RESEARCH published: 08 May 2017 doi: 10.3389/fendo.2017.00099 Leptin Induces Mitosis and Activates the Canonical Wnt/β-Catenin Signaling Pathway in Neurogenic Regions of Xenopus Tadpole Brain Melissa Cui Bender, Christopher J. Sifuentes and Robert J. Denver* Department of Molecular, Cellular and Developmental Biology, University of Michigan, Ann Arbor, MI, USA In addition to its well-known role as an adipostat in adult mammals, leptin has diverse physiological and developmental actions in vertebrates. Leptin has been shown to promote development of hypothalamic circuits and to induce mitosis in different brain areas of mammals. We investigated the ontogeny of leptin mRNA, leptin actions on cell proliferation in the brain, and gene expression in the preoptic area/hypothalamus of tadpoles of Xenopus laevis. The level of leptin mRNA was low in premetamorphic tadpoles, but increased strongly at the beginning of metamorphosis and peaked at metamorphic climax. This increase in leptin mRNA at the onset of metamorphosis correlated with increased cell proliferation in the neurogenic zones of tadpole brain. We found that intracerebroventricular (i.c.v.) injection of recombinant Xenopus leptin Edited by: Maximilian Michel, (rxLeptin) in premetamorphic tadpoles strongly increased cell proliferation in neurogenic University of Michigan, USA zones throughout the tadpole brain. We conducted gene expression profiling of genes Reviewed by: induced at 2 h following i.c.v. injection of rxLeptin. This analysis identified 2,322 genes Suraj Unniappan, induced and 1,493 genes repressed by rxLeptin. The most enriched Kyoto Encyclopedia University of Saskatchewan, Canada Vance Trudeau, of Genes and Genomes term was the canonical Wnt/β-catenin pathway. -

Role of Leptin in Inflammation and Vice Versa
International Journal of Molecular Sciences Review Role of Leptin in Inflammation and Vice Versa Antonio Pérez-Pérez *, Flora Sánchez-Jiménez, Teresa Vilariño-García and Víctor Sánchez-Margalet * Department of Medical Biochemistry and Molecular Biology, and Immunology, Virgen Macarena University Hospital, University of Seville, 41009 Seville, Spain; [email protected] (F.S.-J.); [email protected] (T.V.-G.) * Correspondence: [email protected] (A.P.-P.); [email protected] (V.S.-M.) Received: 16 July 2020; Accepted: 14 August 2020; Published: 16 August 2020 Abstract: Inflammation is an essential immune response for the maintenance of tissue homeostasis. In a general sense, acute and chronic inflammation are different types of adaptive response that are called into action when other homeostatic mechanisms are insufficient. Although considerable progress has been made in understanding the cellular and molecular events that are involved in the acute inflammatory response to infection and tissue injury, the causes and mechanisms of systemic chronic inflammation are much less known. The pathogenic capacity of this type of inflammation is puzzling and represents a common link of the multifactorial diseases, such as cardiovascular diseases and type 2 diabetes. In recent years, interest has been raised by the discovery of novel mediators of inflammation, such as microRNAs and adipokines, with different effects on target tissues. In the present review, we discuss the data emerged from research of leptin in obesity as an inflammatory mediator sustaining multifactorial diseases and how this knowledge could be instrumental in the design of leptin-based manipulation strategies to help restoration of abnormal immune responses. On the other direction, chronic inflammation, either from autoimmune or infectious diseases, or impaired microbiota (dysbiosis) may impair the leptin response inducing resistance to the weight control, and therefore it may be a cause of obesity. -

Leptin Signaling Maintains B-Cell Homeostasis Via Induction of Bcl-2 and Cyclin D1
Leptin signaling maintains B-cell homeostasis via induction of Bcl-2 and Cyclin D1 Queenie Lai Kwan Lama,1, Shengjun Wangb, Otis King Hung Koa, Paul W. Kincadec, and Liwei Lua,1 aDepartment of Pathology and Centre of Infection and Immunology, University of Hong Kong, Hong Kong, China; bDepartment of Immunology, School of Medical Science and Laboratory Medicine, Jiangsu University, Zhenjiang, China; and cImmunobiology and Cancer Program, Oklahoma Medical Research Foundation, Oklahoma City, OK 73104 Edited by Tak Wah Mak, Campbell Family Institute for Breast Cancer Research, Ontario Cancer Institute at Princess Margaret Hospital, University Health Network, Toronto, ON, Canada, and approved June 22, 2010 (received for review April 2, 2010) Regulation of apoptosis and cell cycle progression plays an essential of lymphocytes with impaired humoral responses, but many ques- role in the maintenance of B-cell homeostasis, because a fine tions remain about the molecular mechanisms (13, 14). A variety of balance of survival and expansion is critical for preventing lympho- human cancer cells express the leptin receptor and show enhanced cytic disorders. Although remarkable progress in understanding proliferation in response to leptin stimulation (11). Common B-cell development has been achieved, much less is known concern- polymorphisms in the leptin gene or its receptor are linked to the ing niches that are critical to the maintenance of B-cell homeostasis. pathogenesis of various hematological malignancies including non- Leptin has recently been recognized to be important for modulating Hodgkin’s lymphoma (15). Serum leptin level was significantly the immune responses, but it has remained unclear how leptin sig- elevated in patients with multiple myeloma and chronic lympho- naling influences B-cell physiology.