Association of Phosphatidylinositol-Specific
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Overcalcified Forms of the Coccolithophore Emiliania Huxleyi in High CO2 Waters Are Not Pre-Adapted to Ocean Acidification
Biogeosciences Discuss., https://doi.org/10.5194/bg-2017-303 Manuscript under review for journal Biogeosciences Discussion started: 6 September 2017 c Author(s) 2017. CC BY 4.0 License. Overcalcified forms of the coccolithophore Emiliania huxleyi in high CO2 waters are not pre-adapted to ocean acidification. Peter von Dassow1,2,3*, Francisco Díaz-Rosas1,2, El Mahdi Bendif4, Juan-Diego Gaitán-Espitia5, Daniella Mella-Flores1, Sebastian Rokitta6, Uwe John6, and Rodrigo Torres7,8 5 1 Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago, Chile. 2 Instituto Milenio de Oceanografía de Chile. 3 UMI 3614, Evolutionary Biology and Ecology of Algae, CNRS-UPMC Sorbonne Universités, PUCCh, UACH. 4 Department of Plant Sciences, University of Oxford, OX1 3RB Oxford, UK. 5 CSIRO Oceans and Atmosphere, GPO Box 1538, Hobart 7001, TAS, Australia. 10 6 Marine Biogeosciences | PhytoChange Alfred Wegener Institute – Helmholtz Centre for Polar and Marine Research, Bremerhaven, Germany. 7 Centro de Investigación en Ecosistemas de la Patagonia (CIEP), Coyhaique, Chile. 8 Centro de Investigación: Dinámica de Ecosistemas marinos de Altas Latitudes (IDEAL), Punta Arenas, Chile. Correspondence to: Peter von Dassow ([email protected]) 15 Abstract. Marine multicellular organisms inhabiting waters with natural high fluctuations in pH appear more tolerant to acidification than conspecifics occurring in nearby stable waters, suggesting that environments of fluctuating pH hold genetic reservoirs for adaptation of key groups to ocean acidification (OA). The abundant and cosmopolitan calcifying phytoplankton Emiliania huxleyi exhibits a range of morphotypes with varying degrees of coccolith mineralization. We show that E. huxleyi populations in the naturally acidified upwelling waters of the Eastern South Pacific, where pH drops below 20 7.8 as is predicted for the global surface ocean by the year 2100, are dominated by exceptionally overcalcified morphotypes whose distal coccolith shield can be almost solid calcite. -

Phytoplankton As Key Mediators of the Biological Carbon Pump: Their Responses to a Changing Climate
sustainability Review Phytoplankton as Key Mediators of the Biological Carbon Pump: Their Responses to a Changing Climate Samarpita Basu * ID and Katherine R. M. Mackey Earth System Science, University of California Irvine, Irvine, CA 92697, USA; [email protected] * Correspondence: [email protected] Received: 7 January 2018; Accepted: 12 March 2018; Published: 19 March 2018 Abstract: The world’s oceans are a major sink for atmospheric carbon dioxide (CO2). The biological carbon pump plays a vital role in the net transfer of CO2 from the atmosphere to the oceans and then to the sediments, subsequently maintaining atmospheric CO2 at significantly lower levels than would be the case if it did not exist. The efficiency of the biological pump is a function of phytoplankton physiology and community structure, which are in turn governed by the physical and chemical conditions of the ocean. However, only a few studies have focused on the importance of phytoplankton community structure to the biological pump. Because global change is expected to influence carbon and nutrient availability, temperature and light (via stratification), an improved understanding of how phytoplankton community size structure will respond in the future is required to gain insight into the biological pump and the ability of the ocean to act as a long-term sink for atmospheric CO2. This review article aims to explore the potential impacts of predicted changes in global temperature and the carbonate system on phytoplankton cell size, species and elemental composition, so as to shed light on the ability of the biological pump to sequester carbon in the future ocean. -
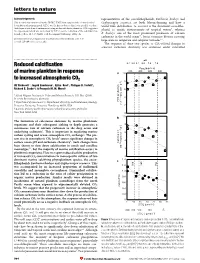
Reduced Calcification of Marine Plankton in Response to Increased
letters to nature Acknowledgements representatives of the coccolithophorids, Emiliania huxleyi and This research was sponsored by the EPSRC. T.W.F. ®rst suggested the electrochemical Gephyrocapsa oceanica, are both bloom-forming and have a deoxidation of titanium metal. G.Z.C. was the ®rst to observe that it was possible to reduce world-wide distribution. G. oceanica is the dominant coccolitho- thick layers of oxide on titanium metal using molten salt electrochemistry. D.J.F. suggested phorid in neritic environments of tropical waters9, whereas the experiment, which was carried out by G.Z.C., on the reduction of the solid titanium dioxide pellets. M. S. P. Shaffer took the original SEM image of Fig. 4a. E. huxleyi, one of the most prominent producers of calcium carbonate in the world ocean10, forms extensive blooms covering Correspondence and requests for materials should be addressed to D. J. F. large areas in temperate and subpolar latitudes9,11. (e-mail: [email protected]). The response of these two species to CO2-related changes in seawater carbonate chemistry was examined under controlled ................................................................. pH Reduced calci®cation 8.4 8.2 8.1 8.0 7.9 7.8 PCO2 (p.p.m.v.) of marine plankton in response 200 400 600 800 a 10 to increased atmospheric CO2 ) 8 –1 Ulf Riebesell *, Ingrid Zondervan*, BjoÈrn Rost*, Philippe D. Tortell², d –1 Richard E. Zeebe*³ & FrancËois M. M. Morel² 6 * Alfred Wegener Institute for Polar and Marine Research, P.O. Box 120161, 4 D-27515 Bremerhaven, Germany mol C cell –13 ² Department of Geosciences & Department of Ecology and Evolutionary Biology, POC production Princeton University, Princeton, New Jersey 08544, USA (10 2 ³ Lamont-Doherty Earth Observatory, Columbia University, Palisades, New York 10964, USA 0 ............................................................................................................................................. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Thioesterase Superfamily Member 1 Undergoes Stimulus-Coupled Conformational Reorganization to Regulate Metabolism in Mice
ARTICLE https://doi.org/10.1038/s41467-021-23595-x OPEN Thioesterase superfamily member 1 undergoes stimulus-coupled conformational reorganization to regulate metabolism in mice Yue Li 1,2, Norihiro Imai3, Hayley T. Nicholls 3, Blaine R. Roberts 4, Samaksh Goyal 1,2, Tibor I. Krisko 3, Lay-Hong Ang 1,2, Matthew C. Tillman4, Anne M. Roberts4, Mahnoor Baqai 1,2, Eric A. Ortlund 4, ✉ ✉ David E. Cohen 3 & Susan J. Hagen 1,2 1234567890():,; In brown adipose tissue, thermogenesis is suppressed by thioesterase superfamily member 1 (Them1), a long chain fatty acyl-CoA thioesterase. Them1 is highly upregulated by cold ambient temperature, where it reduces fatty acid availability and limits thermogenesis. Here, we show that Them1 regulates metabolism by undergoing conformational changes in response to β-adrenergic stimulation that alter Them1 intracellular distribution. Them1 forms metabolically active puncta near lipid droplets and mitochondria. Upon stimulation, Them1 is phosphorylated at the N-terminus, inhibiting puncta formation and activity and resulting in a diffuse intracellular localization. We show by correlative light and electron microscopy that Them1 puncta are biomolecular condensates that are inhibited by phosphorylation. Thus, Them1 forms intracellular biomolecular condensates that limit fatty acid oxidation and sup- press thermogenesis. During a period of energy demand, the condensates are disrupted by phosphorylation to allow for maximal thermogenesis. The stimulus-coupled reorganization of Them1 provides fine-tuning of thermogenesis and energy expenditure. 1 Division of General Surgery, Department of Surgery, Beth Israel Deaconess Medical Center, Boston, MA, USA. 2 Department of Surgery, Harvard Medical School, Boston, MA, USA. 3 Division of Gastroenterology and Hepatology, Department of Medicine, Weill Cornell Medical College, New York, NY, USA. -

Stromatolites Below the Photic Zone in the Northern Arabian Sea Formed by Calcifying Chemotrophic Microbial Mats
Stromatolites below the photic zone in the northern Arabian Sea formed by calcifying chemotrophic microbial mats Tobias Himmler1*, Daniel Smrzka2, Jennifer Zwicker2, Sabine Kasten3,4, Russell S. Shapiro5, Gerhard Bohrmann1, and Jörn Peckmann2,6 1MARUM–Zentrum für Marine Umweltwissenschaften und Fachbereich Geowissenschaften, Universität Bremen, 28334 Bremen, Germany 2Department für Geodynamik und Sedimentologie, Erdwissenschaftliches Zentrum, Universität Wien, 1090 Wien, Austria 3Alfred-Wegener-Institut Helmholtz-Zentrum für Polar- und Meeresforschung, 27570 Bremerhaven, Germany 4Fachbereich Geowissenschaften, Universität Bremen, 28359 Bremen, Germany 5Geological and Environmental Sciences Department, California State University–Chico, Chico, California 95929, USA 6Institut für Geologie, Centrum für Erdsystemforschung und Nachhaltigkeit, Universität Hamburg, 20146 Hamburg, Germany − − + 2− + ABSTRACT reduction: HS + NO3 + H + H2O → SO4 + NH4 (cf. Fossing et al., 1995; Chemosynthesis increases alkalinity and facilitates stromatolite Otte et al., 1999). The effect of this process, referred to as nitrate-driven growth at methane seeps in 731 m water depth within the oxygen sulfide oxidation (ND-SO), is amplified when it takes place near hotspots minimum zone (OMZ) in the northern Arabian Sea. Microbial fab- of SD-AOM (Siegert et al., 2013). Therefore, it has been hypothesized rics, including mineralized filament bundles resembling the sulfide- that fossilization of sulfide-oxidizing bacteria may occur during seep- oxidizing bacterium Thioploca, mineralized extracellular polymeric carbonate formation (Bailey et al., 2009). Yet, seep carbonates resulting substances, and fossilized rod-shaped and filamentous cells, all pre- from the putative interaction of sulfide-oxidizing bacteria with the SD- served in 13C-depleted authigenic carbonate, suggest that biofilm cal- AOM consortium have, to the best of our knowledge, only been recognized cification resulted from nitrate-driven sulfide oxidation (ND-SO) and in ancient seep deposits (Peckmann et al., 2004). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Historical Painting Techniques, Materials, and Studio Practice
Historical Painting Techniques, Materials, and Studio Practice PUBLICATIONS COORDINATION: Dinah Berland EDITING & PRODUCTION COORDINATION: Corinne Lightweaver EDITORIAL CONSULTATION: Jo Hill COVER DESIGN: Jackie Gallagher-Lange PRODUCTION & PRINTING: Allen Press, Inc., Lawrence, Kansas SYMPOSIUM ORGANIZERS: Erma Hermens, Art History Institute of the University of Leiden Marja Peek, Central Research Laboratory for Objects of Art and Science, Amsterdam © 1995 by The J. Paul Getty Trust All rights reserved Printed in the United States of America ISBN 0-89236-322-3 The Getty Conservation Institute is committed to the preservation of cultural heritage worldwide. The Institute seeks to advance scientiRc knowledge and professional practice and to raise public awareness of conservation. Through research, training, documentation, exchange of information, and ReId projects, the Institute addresses issues related to the conservation of museum objects and archival collections, archaeological monuments and sites, and historic bUildings and cities. The Institute is an operating program of the J. Paul Getty Trust. COVER ILLUSTRATION Gherardo Cibo, "Colchico," folio 17r of Herbarium, ca. 1570. Courtesy of the British Library. FRONTISPIECE Detail from Jan Baptiste Collaert, Color Olivi, 1566-1628. After Johannes Stradanus. Courtesy of the Rijksmuseum-Stichting, Amsterdam. Library of Congress Cataloguing-in-Publication Data Historical painting techniques, materials, and studio practice : preprints of a symposium [held at] University of Leiden, the Netherlands, 26-29 June 1995/ edited by Arie Wallert, Erma Hermens, and Marja Peek. p. cm. Includes bibliographical references. ISBN 0-89236-322-3 (pbk.) 1. Painting-Techniques-Congresses. 2. Artists' materials- -Congresses. 3. Polychromy-Congresses. I. Wallert, Arie, 1950- II. Hermens, Erma, 1958- . III. Peek, Marja, 1961- ND1500.H57 1995 751' .09-dc20 95-9805 CIP Second printing 1996 iv Contents vii Foreword viii Preface 1 Leslie A. -

Inflammatory Stimuli Induce Acyl-Coa Thioesterase 7 and Remodeling of Phospholipids Containing Unsaturated Long (C20)-Acyl Chains in Macrophages
Supplemental Material can be found at: http://www.jlr.org/content/suppl/2017/04/17/jlr.M076489.DC1 .html Inflammatory stimuli induce acyl-CoA thioesterase 7 and remodeling of phospholipids containing unsaturated long (C20)-acyl chains in macrophages Valerie Z. Wall,*,† Shelley Barnhart,* Farah Kramer,* Jenny E. Kanter,* Anuradha Vivekanandan-Giri,§ Subramaniam Pennathur,§ Chiara Bolego,** Jessica M. Ellis,§§,*** Miguel A. Gijón,††† Michael J. Wolfgang,*** and Karin E. Bornfeldt1,*,† Department of Medicine,* Division of Metabolism, Endocrinology and Nutrition, and Department of Pathology,† UW Medicine Diabetes Institute, University of Washington, Seattle, WA; Department of Internal Medicine,§ University of Michigan, Ann Arbor, MI; Department of Pharmaceutical and Pharmacological Sciences,** University of Padova, Padova, Italy; Department of Nutrition Science,§§ Purdue University, West Lafayette, IN; Department of Biological Chemistry,*** Johns Hopkins University School of Medicine, Baltimore, MD; and Department of Pharmacology,††† University of Downloaded from Colorado Denver, Aurora, CO Abstract Acyl-CoA thioesterase 7 (ACOT7) is an intracel- containing unsaturated long (C20)-acyl chains in macro- lular enzyme that converts acyl-CoAs to FFAs. ACOT7 is in- phages, and, although ACOT7 has preferential thioesterase duced by lipopolysaccharide (LPS); thus, we investigated activity toward these lipid species, loss of ACOT7 has no ma- www.jlr.org downstream effects of LPS-induced induction of ACOT7 jor detrimental effect on macrophage inflammatory pheno- and its role in inflammatory settings in myeloid cells. Enzy- types.—Wall, V. Z., S. Barnhart, F. Kramer, J. E. Kanter, matic thioesterase activity assays in WT and ACOT7-deficient A. Vivekanandan-Giri, S. Pennathur, C. Bolego, J. M. Ellis, macrophage lysates indicated that endogenous ACOT7 con- M. -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

DIAGNOSIS and THERAPIES for MUCOPOLYSACCHARIDOSES by Francyne Kubaski a Dissertation Submitted to the Faculty of the University
DIAGNOSIS AND THERAPIES FOR MUCOPOLYSACCHARIDOSES by Francyne Kubaski A dissertation submitted to the Faculty of the University of Delaware in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biological Sciences Spring 2017 © 2017 Francyne Kubaski All Rights Reserved DIAGNOSIS AND THERAPIES FOR MUCOPOLYSACCHARIDOSES by Francyne Kubaski Approved: __________________________________________________________ Robin W. Morgan, Ph.D. Chair of the Department of Biological Sciences Approved: __________________________________________________________ George H. Watson, Ph.D. Dean of the College of Arts and Sciences Approved: __________________________________________________________ Ann L. Ardis, Ph.D. Senior Vice Provost for Graduate and Professional Education I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy. Signed: __________________________________________________________ Erica M. Selva, Ph.D. Professor in charge of dissertation I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy. Signed: __________________________________________________________ Shunji Tomatsu, Ph.D. Member of dissertation committee I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy. Signed: __________________________________________________________ Robert W. Mason, Ph.D. Member of dissertation committee I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy.