Construction, Analysis, and Modeling of Complex Reaction Systems Using
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mercury 2.4 User Guide and Tutorials 2011 CSDS Release
Mercury 2.4 User Guide and Tutorials 2011 CSDS Release Copyright © 2010 The Cambridge Crystallographic Data Centre Registered Charity No 800579 Conditions of Use The Cambridge Structural Database System (CSD System) comprising all or some of the following: ConQuest, Quest, PreQuest, Mercury, (Mercury CSD and Materials module of Mercury), VISTA, Mogul, IsoStar, SuperStar, web accessible CSD tools and services, WebCSD, CSD Java sketcher, CSD data file, CSD-UNITY, CSD-MDL, CSD-SDfile, CSD data updates, sub files derived from the foregoing data files, documentation and command procedures (each individually a Component) is a database and copyright work belonging to the Cambridge Crystallographic Data Centre (CCDC) and its licensors and all rights are protected. Use of the CSD System is permitted solely in accordance with a valid Licence of Access Agreement and all Components included are proprietary. When a Component is supplied independently of the CSD System its use is subject to the conditions of the separate licence. All persons accessing the CSD System or its Components should make themselves aware of the conditions contained in the Licence of Access Agreement or the relevant licence. In particular: • The CSD System and its Components are licensed subject to a time limit for use by a specified organisation at a specified location. • The CSD System and its Components are to be treated as confidential and may NOT be disclosed or re- distributed in any form, in whole or in part, to any third party. • Software or data derived from or developed using the CSD System may not be distributed without prior written approval of the CCDC. -

GROMACS: Fast, Flexible, and Free
GROMACS: Fast, Flexible, and Free DAVID VAN DER SPOEL,1 ERIK LINDAHL,2 BERK HESS,3 GERRIT GROENHOF,4 ALAN E. MARK,4 HERMAN J. C. BERENDSEN4 1Department of Cell and Molecular Biology, Uppsala University, Husargatan 3, Box 596, S-75124 Uppsala, Sweden 2Stockholm Bioinformatics Center, SCFAB, Stockholm University, SE-10691 Stockholm, Sweden 3Max-Planck Institut fu¨r Polymerforschung, Ackermannweg 10, D-55128 Mainz, Germany 4Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Nijenborgh 4, NL-9747 AG Groningen, The Netherlands Received 12 February 2005; Accepted 18 March 2005 DOI 10.1002/jcc.20291 Published online in Wiley InterScience (www.interscience.wiley.com). Abstract: This article describes the software suite GROMACS (Groningen MAchine for Chemical Simulation) that was developed at the University of Groningen, The Netherlands, in the early 1990s. The software, written in ANSI C, originates from a parallel hardware project, and is well suited for parallelization on processor clusters. By careful optimization of neighbor searching and of inner loop performance, GROMACS is a very fast program for molecular dynamics simulation. It does not have a force field of its own, but is compatible with GROMOS, OPLS, AMBER, and ENCAD force fields. In addition, it can handle polarizable shell models and flexible constraints. The program is versatile, as force routines can be added by the user, tabulated functions can be specified, and analyses can be easily customized. Nonequilibrium dynamics and free energy determinations are incorporated. Interfaces with popular quantum-chemical packages (MOPAC, GAMES-UK, GAUSSIAN) are provided to perform mixed MM/QM simula- tions. The package includes about 100 utility and analysis programs. -

Package Name Software Description Project
A S T 1 Package Name Software Description Project URL 2 Autoconf An extensible package of M4 macros that produce shell scripts to automatically configure software source code packages https://www.gnu.org/software/autoconf/ 3 Automake www.gnu.org/software/automake 4 Libtool www.gnu.org/software/libtool 5 bamtools BamTools: a C++ API for reading/writing BAM files. https://github.com/pezmaster31/bamtools 6 Biopython (Python module) Biopython is a set of freely available tools for biological computation written in Python by an international team of developers www.biopython.org/ 7 blas The BLAS (Basic Linear Algebra Subprograms) are routines that provide standard building blocks for performing basic vector and matrix operations. http://www.netlib.org/blas/ 8 boost Boost provides free peer-reviewed portable C++ source libraries. http://www.boost.org 9 CMake Cross-platform, open-source build system. CMake is a family of tools designed to build, test and package software http://www.cmake.org/ 10 Cython (Python module) The Cython compiler for writing C extensions for the Python language https://www.python.org/ 11 Doxygen http://www.doxygen.org/ FFmpeg is the leading multimedia framework, able to decode, encode, transcode, mux, demux, stream, filter and play pretty much anything that humans and machines have created. It supports the most obscure ancient formats up to the cutting edge. No matter if they were designed by some standards 12 ffmpeg committee, the community or a corporation. https://www.ffmpeg.org FFTW is a C subroutine library for computing the discrete Fourier transform (DFT) in one or more dimensions, of arbitrary input size, and of both real and 13 fftw complex data (as well as of even/odd data, i.e. -

Chem3d 17.0 User Guide Chem3d 17.0
Chem3D 17.0 User Guide Chem3D 17.0 Table of Contents Recent Additions viii Chapter 1: About Chem3D 1 Additional computational engines 1 Serial numbers and technical support 3 About Chem3D Tutorials 3 Chapter 2: Chem3D Basics 5 Getting around 5 User interface preferences 9 Background settings 10 Sample files 10 Saving to Dropbox 10 Chapter 3: Basic Model Building 12 Default settings 12 Selecting a display mode 12 Using bond tools 13 Using the ChemDraw panel 15 Using other 2D drawing packages 15 Building from text 16 Adding fragments 18 Selecting atoms and bonds 18 Atom charges 21 Object position 23 Substructures 24 Refining models 27 Copying and printing 29 Finding structures online 32 Chapter 4: Displaying Models 35 © Copyright 1998-2017 PerkinElmer Informatics Inc., All rights reserved. ii Chem3D 17.0 Display modes 35 Atom and bond size 37 Displaying dot surfaces 38 Serial numbers 38 Displaying atoms 39 Atom symbols 40 Rotating models 41 Atom and bond properties 44 Showing hydrogen bonds 45 Hydrogens and lone pairs 46 Translating models 47 Scaling models 47 Aligning models 47 Applying color 49 Model Explorer 52 Measuring molecules 59 Comparing models by overlay 62 Molecular surfaces 63 Using stereo pairs 72 Stereo enhancement 72 Setting view focus 73 Chapter 5: Building Advanced Models 74 Dummy bonds and dummy atoms 74 Substructures 75 Bonding by proximity 78 Setting measurements 78 Atom and building types 81 Stereochemistry 85 © Copyright 1998-2017 PerkinElmer Informatics Inc., All rights reserved. iii Chem3D 17.0 Building with Cartesian -

CHEM 743 “Quantum Chemistry” S'15
CHEM 743 \Quantum Chemistry" S'15 Instructor: Sophya Garashchuk∗ Dept of Chemistry and Biochemistry, University of South Carolina, Columbia y (Dated: January 7, 2015) Syllabus • Learning outcomes: (i) the students will gain theoretical knowledge in quantum mechanics and computer skills enabling them to use modern electronic structure codes and simulations with confidence and intelligence to better understand chemical processes; (ii) the students will be able to set up their own input files and interpret/visualize output files for standard quantum chemistry programs/packages that might be relevant to their research; (iii) the students will improve general critical thinking and problem-solving skills as well as improve their skills of independent computer-aided research through extensive use of electronically available resources. • Prerequisites: CHEM 542 or equivalent, i. e. Physical Chemistry - Quantum Mechanics and Spectroscopy • Classes will take place MW 9:40-10:55 AM at PSC 101. We will work with Maple, Q-Chem, Spartan, ADF Class materials and assignments or links to them will be posted on Blackboard Computer and software support: Jun Zhou [email protected] Ph: (803) 777-5492 College of Arts & Sciences Sumwalt College 228 • Office hours: Tue 10-11 am, GSRC 407. I am generally available Mon-Fri 9:00 AM { 5PM. You can just drop by, call or e-mail to check if I am available, or if you have difficulty reaching me, make an appointment. • The suggested general quantum chemistry textbook is \Introduction to Quantum Mechanics in Chemistry", by M. A. Ratner and G. C. Schatz, Prentice-Hall, New Jersey, 2001. Used copies are available through BN.com. -

RMG-Py and Cantherm Documentation ⇌Release 2.0.0
RMG RMG-Py and CanTherm Documentation ⇌Release 2.0.0 William H. Green, Richard H. West, and the RMG Team Sep 16, 2016 CONTENTS 1 RMG User’s Guide 3 1.1 Introduction...............................................3 1.2 Release Notes..............................................4 1.3 Overview of Features...........................................6 1.4 Installation................................................7 1.5 Creating Input Files........................................... 20 1.6 Example Input Files........................................... 29 1.7 Running a Job.............................................. 36 1.8 Analyzing the Output Files........................................ 37 1.9 Species Representation.......................................... 38 1.10 Group Representation.......................................... 39 1.11 Databases................................................. 39 1.12 Thermochemistry Estimation...................................... 58 1.13 Kinetics Estimation........................................... 63 1.14 Liquid Phase Systems.......................................... 65 1.15 Guidelines for Building a Model..................................... 72 1.16 Standalone Modules........................................... 73 1.17 Frequently Asked Questions....................................... 81 1.18 Credits.................................................. 82 1.19 How to Cite................................................ 82 2 CanTherm User’s Guide 83 2.1 Introduction.............................................. -

Pyjac: Analytical Jacobian Generator for Chemical Kinetics
pyJac: analytical Jacobian generator for chemical kinetics Kyle E. Niemeyera,∗, Nicholas J. Curtisb, Chih-Jen Sungb aSchool of Mechanical, Industrial, and Manufacturing Engineering Oregon State University, Corvallis, OR 97331, USA bDepartment of Mechanical Engineering University of Connecticut, Storrs, CT, 06269, USA Abstract Accurate simulations of combustion phenomena require the use of detailed chemical kinet- ics in order to capture limit phenomena such as ignition and extinction as well as predict pollutant formation. However, the chemical kinetic models for hydrocarbon fuels of prac- tical interest typically have large numbers of species and reactions and exhibit high levels of mathematical stiffness in the governing differential equations, particularly for larger fuel molecules. In order to integrate the stiff equations governing chemical kinetics, generally reactive-flow simulations rely on implicit algorithms that require frequent Jacobian matrix evaluations. Some in situ and a posteriori computational diagnostics methods also require accurate Jacobian matrices, including computational singular perturbation and chemical ex- plosive mode analysis. Typically, finite differences numerically approximate these, but for larger chemical kinetic models this poses significant computational demands since the num- ber of chemical source term evaluations scales with the square of species count. Furthermore, existing analytical Jacobian tools do not optimize evaluations or support emerging SIMD processors such as GPUs. Here we introduce pyJac, a Python-based open-source program that generates analytical Jacobian matrices for use in chemical kinetics modeling and anal- ysis. In addition to producing the necessary customized source code for evaluating reaction rates (including all modern reaction rate formulations), the chemical source terms, and the Jacobian matrix, pyJac uses an optimized evaluation order to minimize computational and memory operations. -

Mercury Binding to the Chelation Therapy Agents DMSA and DMPS
Mercury Binding to the Chelation Therapy Agents DMSA and DMPS, and the Rational Design of Custom Chelators for Mercury. Graham N. George,*,† Roger C. Prince,‡ Jürgen Gailer,§ Gavin A. Buttigieg,¶ M. Bonner Denton,¶ Hugh H. Harris⊥ and Ingrid J. Pickering† Department of Geological Sciences, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E2, Canada, ExxonMobil Research and Engineering Company, Annandale, New Jersey 08801, Boehringer Ingelheim Austria GmbH, Dr. Boehringer Gasse 5-11, 1120 Vienna, Austria, Department of Chemistry, The University of Arizona, Tucson, Arizona 85721, School of Chemistry, University of Sydney, Sydney, NSW 2006, Australia. AUTHOR EMAIL ADDRESS: [email protected] RECEIVED DATE (to be automatically inserted after your manuscript is accepted if required according to the journal that you are submitting your paper to) TITLE RUNNING HEAD: Mercury Binding to DMSA and DMPS * To whom correspondence should be addressed at Department of Geological Sciences, University of Saskatchewan, 114 Science Place, Saskatoon, SK, S7N 5E2, Canada. Phone: 306-966-5722, Fax: 306- 966-8593, E-mail: [email protected] † University of Saskatchewan ‡ ExxonMobil Research and Engineering § Boehringer Ingelheim Austria ¶ University of Arizona ⊥ University of Sydney 1 ABSTRACT Clinical chelation therapy of mercury poisoning generally uses one or both of two drugs – meso- dimercaptosuccinic acid (DMSA), and dimercaptopropanesulfonic acid (DMPS), commercially sold as ® ® Chemet and Dimaval , respectively. We have used a combination of mercury LIII-edge X-ray absorption spectroscopy and density functional theory calculations to investigate the chemistry of interaction of mercuric ions with each of these chelation therapy drugs. We show that neither DMSA nor DMPS forms a true chelate complex with mercuric ions, and that these drugs should be considered sub-optimal for their clinical task of binding mercuric ions. -
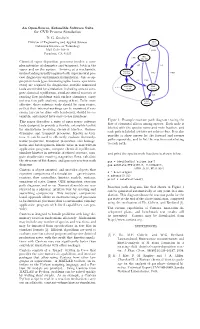
An Open-Source, Extensible Software Suite for CVD Process Simulation D
An Open-Source, Extensible Software Suite for CVD Process Simulation D. G. Goodwin Division of Engineering and Applied Science SIH4 0.0016 California Institute of Technology Mail Code 104-44 0.0135 Pasadena, CA 91125 SIH2 0.852 4.8e-005 Chemical vapor deposition processes involve a com- plex interplay of chemistry and transport, both in the 0.85 0.4 vapor and on the surface. Arriving at a mechanistic H3SISIH 0.415 0.429 understanding usually requires both experimental pro- 2.9e-005 cess diagnostics and numerical simulation. Just as ap- propriate tools (gas chromatographs, lasers, spectrom- 1 0.8 0.429 eters) are required for diagnostics, suitable numerical H2SISIH2 SI3H8 0.101 tools are needed for simulation, including ones to com- 0.0038 7.3e-005 pute chemical equilibrium, simulate stirred reactors or reacting flow problems with surface chemistry, carry 0.83 out reaction path analysis, among others. To be most SI2H6 effective, these software tools should be open-source, 0.00019 so that their internal workings can be examined if nec- essary (as can be done with hardware), should be ex- tensible, and should have easy-to-use interfaces. Figure 1: Example reaction path diagram tracing the This paper describes a suite of open-source software flow of elemental silicon among species. Each node is tools designed to provide a flexible, extensible toolkit labeled with the species name and mole fraction, and for simulations involving chemical kinetics, thermo- each path is labeled with its net relative flux. It is also dynamics, and transport processes. Known as Can- possible to show arrows for the forward and reverse tera, it can be used to efficiently evaluate thermody- paths separately, and to list the reaction contributing namic properties, transport properties, and homoge- to each path. -

Teaching with SCIGRESS
Teaching with SCIGRESS Exercises on Molecular Modeling in Chemistry Crispin Wong and James Currie, eds. Pacific University Teaching with SCIGRES Molecular Modeling in Chemistry Crispin Wong and James Currie Editors Pacific University Forest Grove, Oregon 2 Copyright statement © 2001 - 2012 Fujitsu Limited. All rights reserved. This document may not be used, sold, transferred, copied or reproduced, in any manner or form, or in any medium, to any person other than with the prior written consent of Fujitsu Limited. Published by Fujitsu Limited. All possible care has been taken in the preparation of this publication. Fujitsu Limited does not accept liability for any inaccuracies that may be found. Fujitsu Limited reserves the right to make changes without notice both to this publication and to the product it describes. 3 Table of Contents Exercises 7 Part 1: General Chemistry Experiment 1 Calculating the Geometry of Molecules and Ions 16 Experiment 2 Kinetics of Substitution Reactions 29 Experiment 3 Sunscreen and Ultraviolet Absorption 38 Part 2: Organic Chemistry Experiment 4 Evaluations of Conformations of Menthone and Isomenthone 46 Experiment 5 The Evelyn Effect 52 Part 3: Physical Chemistry Experiment 6 Investigating the Resonance Stabilization of Benzene 63 Experiment 7 Investigation of the Rotational Barrier in N-N-dimethylacetamide 68 Part 4: Inorganic Chemistry Experiment 8 Polarities of Small Molecules 75 Experiment 9 Structures of Molecules Which Exceed the Octet Rule. 77 Experiment 10 Comparison of Gas Phase Acidities of Binary -

BIOVIA MATERIALS STUDIO OVERVIEW Datasheet
BIOVIA MATERIALS STUDIO OVERVIEW Datasheet 10 Å 3 nm 15 µm Modeling and Simulation for Next Generation Materials. BIOVIA Materials Studio® is a complete modeling and simulation environment that enables researchers in materials science and chemistry to develop new materials by predicting the relationships of a material’s atomic and molecular structure with its properties and behavior. Using Materials Studio, researchers in many industries can engineer better performing materials of all types, including pharmaceuticals, catalysts, polymers and composites, metals and alloys, batteries and fuel cells, nanomaterials, and more. Materials Studio is the world’s most advanced, yet easy Materials Visualizer provides capability to construct, manipulate to use environment for modeling and evaluating materials and view models of molecules, crystalline materials, surfaces, performance and behavior. Using Materials Studio, materials polymers, and mesoscale structures. It also supports the full scientists experience the following benefits: range of Materials Studio simulations with capabilities to • Reduction in cost and time associated with physical testing visualize results through images, animations, graphs, charts, and experimentation through “Virtual Screening” of candidate tables, and textual data. Most tools in the Materials Visualizer material variations. can also be accessed through the MaterialsScript API, allowing • Acceleration of the innovation process - developing new, better expert users to create custom capabilities and automate performing, more sustainable, and cost effective materials faster repetitive tasks. Materials Visualizer’s Microsoft Windows than can be done with physical testing and experimentation. client operates with a range of Windows and Linux server • Improved fundamental understanding of the relationship architectures to provide a highly responsive user experience. between atomic and molecular structure with material properties and behavior. -

Quick Navigation Mechanical Engineering Department Facilities
Quick Navigation Department Facilities Multi-User Facilities Other Facilities Mechanical Engineering Department Facilities Departmental Facilities and Individual Faculty Research Laboratories: Shared Services Facilities Director: Haejune Kim We provide students and researchers open access to a wide range of equipment for measuring, characterizing and imaging of samples. If necessary, shared facility staff or super user of the instrument will offer training before giving access to the equipment. The shared services facilities are available to mechanical engineering faculty and students. Currently the group maintains and has available for scheduling the following machines:VEGA II LSU SEM, KLA- Tencor P-6 Stylus Profiler, VHX-600 Digital Microscope, High Performance Liquid Chromatography (HPLC), Total Organic Carbon (TOC) Analyzer, Atomic Layer Deposition (ALD) System, MicroClimate environmental chamber, Photron FSTCAM SA5 High Speed Camera, Olympus BX 61/Visitech QLC-100 Confocal Microscopy, Simultaneous Thermal Analyzer – Q600 SDT, Differential Scanning Calorimeter – DSC Q1000, Veeco Dektak 150 Profilometer, and Salt Spray Chamber. Acoustics and Signal Processing Laboratory Contact: Dr. Yong-Joe Kim The ASPL, founded in 2009, is a research laboratory of the Department of Mechanical Engineering at Texas A&M University (TAMU), College Station, Texas, USA. It is located at James J. Cain ’51 Building #409. Prof. Yong-Joe Kim currently serves as the director for the ASPL. The current research is focused on the areas of acoustics, signal processing, vibration, dynamics, and biomechanics. Advanced Computational Mechanics Laboratory Contact: Dr. J.N. Reddy Professor Reddy’s Advanced Computational Mechanics Laboratory (ACML) at Texas A&M University is dedicated to state-of-the-art research in the development of novel mathematical models and numerical simulation of physical phenomena.