Influence of Maize Mosaic Virus and Host Plant Genotypes on the Bio
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Multiplication of Maize Stripe Virus in Peregrinus Maidis
Vector Relations Multiplication of Maize Stripe Virus in Peregrinusmaidis L. R. Nault and D. T. Gordon Professor, Department of Entomology, and professor, Department of Plant Pathology, The Ohio State University (OSU), Ohio Agricultural Research and Development Center (OARDC), Wooster 44691. Submitted as Journal Article 147-87 by the OSU-OARDC. Salaries and research support were provided by state and federal funds appropriated to the OSU-OARDC. We gratefully acknowledge the technical assistance of Lakshman S. Negi, Marian Coffey, and Amy Juan Rubink in performing the enzyme-linked immunosorbent assays, W. E. Styer for the preparation of planthoppers and planthopper organs for assay, and N. H. Gordon for advice on the statistical analysis of the enzyme-linked immunosorbent assay data. Accepted for publication 26 February 1988. ABSTRACT Nault, L. R., and Gordon, D. T. 1988. Multiplication of maize stripe virus in Peregrinusmaidis. Phytopathology 78:991-995. The enzyme-linked immunosorbent assay (ELISA) was used to bursa copulatrix, and single eggs of viruliferous females; and accessory demonstrate that the maize stripe virus (MStpV) multiplies in its delphacid gland and seminal vesicle of viruliferous males. In this last group, none of planthopper vector, Peregrinusmaidis. ELISAs were performed using an the 10 testes were found to be positive for MStpV by ELISA. In a time antiserum to purified MStpV. MStpV was detected by ELISA from a single course experiment, MStpV was detected by ELISA from the midgut and planthopper immediately after a 2-day acquisition access period (AAP) ovaries but not the salivary glands at 0 and 2 days after a 7-day AAP. -

Maize Stripe Tenuivirus1
Plant Pathology Circular No. 363 Fla. Dept. Agric. & Consumer Services January/February 1994 Division of Plant Industry Maize Stripe Tenuivirus1 J. H. Tsai2 and L. G. Brown3 INTRODUCTION: Maize stripe is a severe viral disease of maize in the southern U. S., Central America, Africa, Australia, and several Pacific islands, and probably occurs in most tropical maize growing regions (Gingery 1985; Tsai and Zitter 1982). The first report of this disease in the Continental U.S. was in Florida in 1975 (Tsai 1975). A serious outbreak of a disease complex in maize occurred in south Florida from 1979-1980. At least five viral and two mollicute (mycoplasmas that lack a true cell wall) plant pathogens were involved in the outbreak. Maize stripe virus (MStV) was considered the most important component in the disease epidemic (Bradfute et al. 1981; Tsai and Falk 1988). SYMPTOMS AND HOST RANGE: Initial symptoms are fine chlorotic stipplings between leaf veins which later develop into continuous chlorotic stripes of varying width and intensity (Tsai 1975) (Fig. 1). Young plants infected at the 4- to 5-leaf stage often exhibit complete chlorosis on the emerging whorl leaf, and the center leaf usually remains folded and bent (Tsai 1975). The host range of MStV includes Zea spp., several Sorghum spp., and itchgrass (Rouboellia exaltata L.f.) (Greber 1983, Tsai and Falk 1988). Itchgrass is a noxious weed which was introduced into southern Florida from tropical Asia (Hitchock and Chase 1951). Figure 1. Maize stripe virus on Zea mays. Fine Chlorotic stipplings (left). Continuous chlorotic stripes (right). DISEASE DEVELOPMENT: MStV is transmitted by the corn delphacid, Peregrinus maidis (Ashmead) (Homoptera: Delphacidae) in a persistent-propagative manner (Fig. -

Rice Hoja Blanca: a Complex Plant–Virus–Vector Pathosystem F.J
CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources 2010 5, No. 043 Review Rice hoja blanca: a complex plant–virus–vector pathosystem F.J. Morales* and P.R. Jennings Address: International Centre for Tropical Agriculture, AA 6713, Cali, Colombia. *Correspondence: F.J. Morales. E-mail: [email protected] Received: 30 April 2010 Accepted: 24 June 2010 doi: 10.1079/PAVSNNR20105043 The electronic version of this article is the definitive one. It is located here: http://www.cabi.org/cabreviews g CAB International 2009 (Online ISSN 1749-8848) Abstract Rice hoja blanca (RHB; white leaf) devastated rice (Oryza sativa) plantings in tropical America for half a century, before scientists could either identify its causal agent or understand the nature of its cyclical epidemics. The association of the planthopper Tagosodes orizicolus with RHB outbreaks, 20 years after its emergence in South America, suggested the existence of a viral pathogen. However, T. orizicolus could also cause severe direct feeding damage (hopperburn) to rice in the absence of hoja blanca, and breeders promptly realized that the genetic basis of resistance to these problems was different. Furthermore, it was observed that the causal agent of RHB could only be trans- mitted by a relatively low proportion of the individuals in any given population of T. orizicolus and that the pathogen was transovarially transmitted to the progeny of the planthopper vectors, affecting their normal biology. An international rice germplasm screening effort was initiated in the late 1950s to identify sources of resistance against RHB and the direct feeding damage caused by T. -

Taxonomy of the Order Bunyavirales: Update 2019
Archives of Virology (2019) 164:1949–1965 https://doi.org/10.1007/s00705-019-04253-6 VIROLOGY DIVISION NEWS Taxonomy of the order Bunyavirales: update 2019 Abulikemu Abudurexiti1 · Scott Adkins2 · Daniela Alioto3 · Sergey V. Alkhovsky4 · Tatjana Avšič‑Županc5 · Matthew J. Ballinger6 · Dennis A. Bente7 · Martin Beer8 · Éric Bergeron9 · Carol D. Blair10 · Thomas Briese11 · Michael J. Buchmeier12 · Felicity J. Burt13 · Charles H. Calisher10 · Chénchén Cháng14 · Rémi N. Charrel15 · Il Ryong Choi16 · J. Christopher S. Clegg17 · Juan Carlos de la Torre18 · Xavier de Lamballerie15 · Fēi Dèng19 · Francesco Di Serio20 · Michele Digiaro21 · Michael A. Drebot22 · Xiaˇoméi Duàn14 · Hideki Ebihara23 · Toufc Elbeaino21 · Koray Ergünay24 · Charles F. Fulhorst7 · Aura R. Garrison25 · George Fú Gāo26 · Jean‑Paul J. Gonzalez27 · Martin H. Groschup28 · Stephan Günther29 · Anne‑Lise Haenni30 · Roy A. Hall31 · Jussi Hepojoki32,33 · Roger Hewson34 · Zhìhóng Hú19 · Holly R. Hughes35 · Miranda Gilda Jonson36 · Sandra Junglen37,38 · Boris Klempa39 · Jonas Klingström40 · Chūn Kòu14 · Lies Laenen41,42 · Amy J. Lambert35 · Stanley A. Langevin43 · Dan Liu44 · Igor S. Lukashevich45 · Tāo Luò1 · Chuánwèi Lüˇ 19 · Piet Maes41 · William Marciel de Souza46 · Marco Marklewitz37,38 · Giovanni P. Martelli47 · Keita Matsuno48,49 · Nicole Mielke‑Ehret50 · Maria Minutolo3 · Ali Mirazimi51 · Abulimiti Moming14 · Hans‑Peter Mühlbach50 · Rayapati Naidu52 · Beatriz Navarro20 · Márcio Roberto Teixeira Nunes53 · Gustavo Palacios25 · Anna Papa54 · Alex Pauvolid‑Corrêa55 · Janusz T. Pawęska56,57 · Jié Qiáo19 · Sheli R. Radoshitzky25 · Renato O. Resende58 · Víctor Romanowski59 · Amadou Alpha Sall60 · Maria S. Salvato61 · Takahide Sasaya62 · Shū Shěn19 · Xiǎohóng Shí63 · Yukio Shirako64 · Peter Simmonds65 · Manuela Sironi66 · Jin‑Won Song67 · Jessica R. Spengler9 · Mark D. Stenglein68 · Zhèngyuán Sū19 · Sùróng Sūn14 · Shuāng Táng19 · Massimo Turina69 · Bó Wáng19 · Chéng Wáng1 · Huálín Wáng19 · Jūn Wáng19 · Tàiyún Wèi70 · Anna E. -

A Look Into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins
viruses Review A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins Shanna S. Leventhal, Drew Wilson, Heinz Feldmann and David W. Hawman * Laboratory of Virology, Rocky Mountain Laboratories, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA; [email protected] (S.S.L.); [email protected] (D.W.); [email protected] (H.F.) * Correspondence: [email protected]; Tel.: +1-406-802-6120 Abstract: In 2016, the Bunyavirales order was established by the International Committee on Taxon- omy of Viruses (ICTV) to incorporate the increasing number of related viruses across 13 viral families. While diverse, four of the families (Peribunyaviridae, Nairoviridae, Hantaviridae, and Phenuiviridae) contain known human pathogens and share a similar tri-segmented, negative-sense RNA genomic organization. In addition to the nucleoprotein and envelope glycoproteins encoded by the small and medium segments, respectively, many of the viruses in these families also encode for non-structural (NS) NSs and NSm proteins. The NSs of Phenuiviridae is the most extensively studied as a host interferon antagonist, functioning through a variety of mechanisms seen throughout the other three families. In addition, functions impacting cellular apoptosis, chromatin organization, and transcrip- tional activities, to name a few, are possessed by NSs across the families. Peribunyaviridae, Nairoviridae, and Phenuiviridae also encode an NSm, although less extensively studied than NSs, that has roles in antagonizing immune responses, promoting viral assembly and infectivity, and even maintenance of infection in host mosquito vectors. Overall, the similar and divergent roles of NS proteins of these Citation: Leventhal, S.S.; Wilson, D.; human pathogenic Bunyavirales are of particular interest in understanding disease progression, viral Feldmann, H.; Hawman, D.W. -

HTS-Based Diagnostics of Sugarcane Viruses: Seasonal Variation and Its Implications for Accurate Detection
viruses Article HTS-Based Diagnostics of Sugarcane Viruses: Seasonal Variation and Its Implications for Accurate Detection Martha Malapi-Wight 1,*,† , Bishwo Adhikari 1 , Jing Zhou 2, Leticia Hendrickson 1, Clarissa J. Maroon-Lango 3, Clint McFarland 4, Joseph A. Foster 1 and Oscar P. Hurtado-Gonzales 1,* 1 USDA-APHIS Plant Germplasm Quarantine Program, Beltsville, MD 20705, USA; [email protected] (B.A.); [email protected] (L.H.); [email protected] (J.A.F.) 2 Department of Agriculture, Agribusiness, Environmental Sciences, Texas A&M University-Kingsville, Kingsville, TX 78363, USA; [email protected] 3 USDA-APHIS-PPQ-Emergency and Domestic Program, Riverdale, MD 20737, USA; [email protected] 4 USDA-APHIS-PPQ-Field Operations, Raleigh, NC 27606, USA; [email protected] * Correspondence: [email protected] (M.M.-W.); [email protected] (O.P.H.-G.) † Current affiliation: USDA-APHIS BRS Biotechnology Risk Analysis Program, Riverdale, MD 20737, USA. Abstract: Rapid global germplasm trade has increased concern about the spread of plant pathogens and pests across borders that could become established, affecting agriculture and environment systems. Viral pathogens are of particular concern due to their difficulty to control once established. A comprehensive diagnostic platform that accurately detects both known and unknown virus species, as well as unreported variants, is playing a pivotal role across plant germplasm quarantine programs. Here we propose the addition of high-throughput sequencing (HTS) from total RNA to the routine Citation: Malapi-Wight, M.; quarantine diagnostic workflow of sugarcane viruses. We evaluated the impact of sequencing depth Adhikari, B.; Zhou, J.; Hendrickson, needed for the HTS-based identification of seven regulated sugarcane RNA/DNA viruses across L.; Maroon-Lango, C.J.; McFarland, two different growing seasons (spring and fall). -

Rice Fact Sheet
Rice Grassy Stunt Rice Grassy Stunt – Page 1 of 3 Known Distribution Rice grassy stunt virus (RGSV) disease (grassy stunt) occurs in South and Southeast Asia, China, Japan, and Taiwan (Hibino 1996). RGSV incidence was high from 1970 to 1977 in Indonesia; from 1973 to 1977 and from 1982 to 1983 in the Philippines (Hibino 1989, Hibino et al 1985); from 1973 to 1974 and 1981 in Kerala, India (Kulshreshtha et al 1974); in 1972 and 1984 in Tamil Nadu, India (Mariappan et al 1984); and in 1978 in Kyushu, Japan (Iwasaki and Shinkai 1979). High incidences of grassy stunt together with ragged stunt disease (ragged stunt) were also reported in the Mekong Delta of Vietnam during 2000 to 2007 (Du et al 2005, Huan and Heong 2000). Causal Virus Rice grassy stunt virus (Fig. 1) is a member of the Tenuivirus group, which consists of 6 members along with rice stripe virus as the type species (Hibino 1986, Toriyama 1995, Mayo et al 2000, Toriyama s 2004). The RGSV particle is a circular filament of 6 to 8 nm in width. t The filamentous particles of RGSV are ribonucleoproteins, which are composed of a single nucleocapsid protein and genomic single- stranded RNA segments (Ramirez and Haenni 1994). RGSV differs from other tenuiviruses in that it possesses 6 ambisense RNA segments (Miranda et al 2000, Toriyama et al 1997, 1998). RNAs 1, 2, 5, and 6 are equivalent to rice stripe virus (RSV) RNAs 1, 2, 3, and 4, respectively, and RGSV RNAs 3 and 4 are unique in RGSV. -

2008.021P 2008.022P 2008.023P
Taxonomic proposal to the ICTV Executive Committee This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). Code(s) assigned: 2008.021-025P (to be completed by ICTV officers) Short title: Creation of genus Emaravirus for the new species European mountain ash ringspot-associated virus (e.g. 6 new species in the genus Zetavirus; re-classification of the family Zetaviridae etc.) Modules attached 1 2 3 4 5 (please check all that apply): 6 7 Author(s) with e-mail address(es) of the proposer: Nicole Mielke, [email protected] Nanette Schlatermund, [email protected] Hans-Peter Muehlbach, [email protected] ICTV-EC or Study Group comments and response of the proposer: MODULE 4: NEW GENUS (if more than one genus is to be created, please complete additional copies of this section) Code 2008.021P (assigned by ICTV officers) To create a new genus assigned as follows: Subfamily: - Fill in all that apply. Ideally, a genus should be placed within a higher taxon, Family: Unassigned (ssRNA –ve) but if not put “unassigned” here. Order: - Code 2008.022P (assigned by ICTV officers) To name the new genus: Emaravirus Code 2008.023P (assigned by ICTV officers) To assign the following as species in the new genus: You may list several species here. For each species, please state whether it is new or existing. If the species is new, please complete Module 5 to create it. If the species already exists, please state whether it is unassigned or is to be removed from another genus and, if the latter, complete module 6(a) to ‘REMOVE’ it from that genus. -

Downloaded and De Novo Assembled Using CLC
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.11.245274; this version posted August 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Ever-increasing viral diversity associated with the red imported fire ant Solenopsis invicta 2 (Formicidae: Hymenoptera) 3 4 César A.D. Xavier1, Margaret L. Allen2*, and Anna E. Whitfield1* 5 6 1Department of Entomology and Plant Pathology, North Carolina State University, 840 Main Campus 7 Drive, Raleigh, NC 27606, USA 8 2U. S. Department of Agriculture, Agricultural Research Service, Biological Control of Pests Research 9 Unit, 59 Lee Road, Stoneville, MS 38776, USA 10 *Corresponding author: Margaret L. Allen 11 Phone: 1(662) 686-3647 12 E-mail: [email protected] 13 *Corresponding author: Anna E. Whitfield 14 Phone: 1(919) 515-5861 15 E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.11.245274; this version posted August 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 16 Abstract 17 Background: Advances in sequencing and analysis tools have facilitated discovery of many new 18 viruses from invertebrates, including ants. Solenopsis invicta is an invasive ant that has quickly 19 spread around world causing significant ecological and economic impacts. Its virome has begun 20 to be characterized pertaining to potential use of viruses as natural enemies. Although the S. invicta 21 virome is best characterized among ants, most studies have been performed in its native range, 22 with little information from invaded areas. -

Tsai: Biology of Peregrinus Maidis 19 DEVELOPMENT AND
Tsai: Biology of Peregrinus maidis 19 DEVELOPMENT AND OVIPOSITION OF PEREGRINUS MAIDIS (HOMOPTERA: DELPHACIDAE) ON VARIOUS HOST PLANTS JAMES H. TSAI Fort Lauderdale Research and Education Center University of Florida, IFAS Fort Lauderdale, FL 33314 ABSTRACT The development and oviposition of Peregrinus maidis (Ashmead) (Homoptera: Delphacidae), a serious pest and the only known vector of maize stripe tenuivirus and maize mosaic rhabdovirus in tropical and subtropical areas, was studied on the fol- lowing plants in the laboratory: corn (Zea mays L. var. Saccharata ‘Guardian’), itch grass (Rottboellia exaltata L.), rice (Oryza sativa L. var. Mars, Saturn, Nato, Bellevue, Labelle, Labonnet, and Starbonnet), sorghum (Sorghum bicolor (L.) Moench var. AKS 614), goose grass (Eleusine indica (L.) Gaertn), oats (Avena sativa L.), rye (Secale ce- reale L.), gama grass (Tripsacum dactyloides L.), barnyard grass (Echinochloa crus- galli L.) and sugarcane (Saccharum officinarum L.). Peregrinus maidis nymphs did 20 Florida Entomologist 79(1) March, 1996 not develop on rye, oats, rice and sugarcane, but the adults survived for various lengths of time on these test plants. The average length of nymphal development on corn, itch grass, sorghum, goose grass, barnyard grass and gama grass was 17.20, 17.87, 20.21, 24.97, 27.24 and 60.50 days, respectively. Adult longevity (X ± SD) on corn, gama grass, itch grass, sorghum, goose grass, and barnyard grass was 36.1 ± 20.0, 42.7 ± 16.6, 28.3 ± 11.9, 7.6 ± 6.4, 8.1 ± 7.3 and 7.3 ± 6.6 days, respectively. Ovi- position rarely occurred on sorghum, goose grass and barnyard grass. -
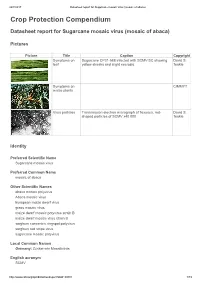
Datasheet Report for Sugarcane Mosaic Virus (Mosaic of Abaca)
24/7/2017 Datasheet report for Sugarcane mosaic virus (mosaic of abaca) Crop Protection Compendium Datasheet report for Sugarcane mosaic virus (mosaic of abaca) Pictures Picture Title Caption Copyright Symptoms on Sugarcane CP31-588 infected with SCMV-SC showing David S. leaf yellow streaks and slight necrosis. Teakle Symptoms on CIMMYT maize plants Virus particles Transmission electron micrograph of flexuous, rod- David S. shaped particles of SCMV x40 000. Teakle Identity Preferred Scientific Name Sugarcane mosaic virus Preferred Common Name mosaic of abaca Other Scientific Names abaca mosaic potyvirus Abaca mosaic virus European maize dwarf virus grass mosaic virus maize dwarf mosaic potyvirus strain B maize dwarf mosaic virus strain B sorghum concentric ringspot potyvirus sorghum red stripe virus sugarcane mosaic potyvirus Local Common Names Germany: Zuckerrohr Mosaikvirus English acronym SCMV http://www.cabi.org/cpc/datasheetreport?dsid=49801 1/19 24/7/2017 Datasheet report for Sugarcane mosaic virus (mosaic of abaca) EPPO code SCMV00 (Sugarcane mosaic potyvirus) Taxonomic Tree Domain: Virus Group: "Positive sense ssRNA viruses" Group: "RNA viruses" Family: Potyviridae Genus: Potyvirus Species: Sugarcane mosaic virus Notes on Taxonomy and Nomenclature This virus was described from sugarcane by Brandes (1919). Strains naturally infecting certain other hosts are also known as abaca mosaic potyvirus (Eloja and Tinsley, 1963) and as maize dwarf mosaic potyvirus strain B (Mackenzie et al., 1966; Louie and Knoke, 1975). Until recently, most aphid-borne potyviruses infecting sugarcane and other members of the Poaceae were assumed to be either sugarcane mosaic virus or maize dwarf mosaic virus. However, new criteria for speciation of these members of the Potyviridae have been proposed (Shukla et al., 1989; Shukla et al., 1992; Shukla and Ward, 1994): the amino-acid sequence homology of their coat proteins, and serological relationships with antisera derived from epitopes in the amino terminus rather than the core region of the coat protein. -

Virus Diseases and Crop Biosecurity
Virus Diseases and Crop Biosecurity NATO Security through Science Series This Series presents the results of scientific meetings supported under the NATO Programme for Security through Science (STS) Meetings supported by the NATO STS Programme are in security-related priority areas of Defence Against Terrorism or Countering Other Threats to Security. The types of meeting supported are generally “Advanced Study Institute” and “Advanced Research Workshops”. The NATO STS Series collects together the results of these meetings. The meetings are co-organized by scientist from NATO countries and scientists from NATO’s “Partner” or “Mediterranean Dialogue” countries. The observations and recommendations made at the meetings, as well as the contents of the volumes in the Series, reflect those of participants and contributors only; they should not necessarily be regarded as reflecting NATO views or policy. Advanced Study Institutes (ASI) are high-level tutorial courses to convey the latest developments in a subject to an advanced-level audience Advanced Research Workshops (ARW) are expert meetings where an intense but informal exchange of views at the frontiers of a subject aims at identifying directions for future actions Following a transformation of the programme in 2004 the Series has been re-named and re-organised. Recent volumes on topics not related to security, which result from meetings supported under the programme earlier, may be found in the NATO Science Series. The Series is published by IOS Press, Amsterdam, and Springer, Dordrecht, in conjunction with the NATO Public Diplomacy Division. Sub-Series A. Chemistry and Biology Springer B. Physics and Biophysics Springer C. Environmental Security Springer D.