Beyond Fish Edna Metabarcoding: Field Replicates Disproportionately Improve the Detection of Stream Associated Vertebrate Specie
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Importance of Muraviovka Park, Amur Province, Far East Russia, For
FORKTAIL 33 (2017): 81–87 The importance of Muraviovka Park, Amur province, Far East Russia, for bird species threatened at regional, national and international level based on observations between 2011 and 2016 WIELAND HEIM & SERGEI M. SMIRENSKI The middle reaches of the Amur River in Far East Russia are still an under-surveyed region, yet holding a very high regional biodiversity. During a six-year survey at Muraviovka Park, a non-governmental nature reserve, 271 bird species have been recorded, 14 of which are globally threatened, highlighting the importance of this area for bird conservation. INTRODUCTION RESULTS Recent studies have shown that East Asia and especially the Amur A total of 271 species was recorded inside Muraviovka Park between basin hold huge numbers of endangered species, and the region was 2011 and 2016; 24 species are listed as Near Treatened (NT), designated as a hotspot of threatened biodiversity (e.g. Vignieri 2014). Vulnerable (VU), Endangered (EN) or Critically Endangered (CR) Tis is especially true for birds. Te East Asian–Australasian Flyway (BirdLife International 2017a), 31 species in the Russian Red Data is not only one of the richest in species and individuals but is also the Book (Iliashenko & Iliashenko 2000) (Ru) and 60 species in the least surveyed and most threatened fyway (Yong et al. 2015). Current Amur region Red Data Book (Glushchenko et al. 2009) (Am). In data about distribution, population size and phenology are virtually the case of the Russian and Amur regional Red Data Books, the lacking for many regions, including the Amur region, Far East Russia. -

Review and Meta-Analysis of the Environmental Biology and Potential Invasiveness of a Poorly-Studied Cyprinid, the Ide Leuciscus Idus
REVIEWS IN FISHERIES SCIENCE & AQUACULTURE https://doi.org/10.1080/23308249.2020.1822280 REVIEW Review and Meta-Analysis of the Environmental Biology and Potential Invasiveness of a Poorly-Studied Cyprinid, the Ide Leuciscus idus Mehis Rohtlaa,b, Lorenzo Vilizzic, Vladimır Kovacd, David Almeidae, Bernice Brewsterf, J. Robert Brittong, Łukasz Głowackic, Michael J. Godardh,i, Ruth Kirkf, Sarah Nienhuisj, Karin H. Olssonh,k, Jan Simonsenl, Michał E. Skora m, Saulius Stakenas_ n, Ali Serhan Tarkanc,o, Nildeniz Topo, Hugo Verreyckenp, Grzegorz ZieRbac, and Gordon H. Coppc,h,q aEstonian Marine Institute, University of Tartu, Tartu, Estonia; bInstitute of Marine Research, Austevoll Research Station, Storebø, Norway; cDepartment of Ecology and Vertebrate Zoology, Faculty of Biology and Environmental Protection, University of Lodz, Łod z, Poland; dDepartment of Ecology, Faculty of Natural Sciences, Comenius University, Bratislava, Slovakia; eDepartment of Basic Medical Sciences, USP-CEU University, Madrid, Spain; fMolecular Parasitology Laboratory, School of Life Sciences, Pharmacy and Chemistry, Kingston University, Kingston-upon-Thames, Surrey, UK; gDepartment of Life and Environmental Sciences, Bournemouth University, Dorset, UK; hCentre for Environment, Fisheries & Aquaculture Science, Lowestoft, Suffolk, UK; iAECOM, Kitchener, Ontario, Canada; jOntario Ministry of Natural Resources and Forestry, Peterborough, Ontario, Canada; kDepartment of Zoology, Tel Aviv University and Inter-University Institute for Marine Sciences in Eilat, Tel Aviv, -
Labidesthes Sicculus
Version 2, 2015 United States Fish and Wildlife Service Lower Great Lakes Fish and Wildlife Conservation Office 1 Atherinidae Atherinidae Sand Smelt Distinguishing Features: — (Atherina boyeri) — Sand Smelt (Non-native) Old World Silversides Old World Silversides Old World (Atherina boyeri) Two widely separated dorsal fins Eye wider than Silver color snout length 39-49 lateral line scales 2 anal spines, 13-15.5 rays Rainbow Smelt (Non -Native) (Osmerus mordax) No dorsal spines Pale green dorsally Single dorsal with adipose fin Coloring: Silver Elongated, pointed snout No anal spines Size: Length: up to 145mm SL Pink/purple/blue iridescence on sides Distinguishing Features: Dorsal spines (total): 7-10 Brook Silverside (Native) 1 spine, 10-11 rays Dorsal soft rays (total): 8-16 (Labidesthes sicculus) 4 spines Anal spines: 2 Anal soft rays: 13-15.5 Eye diameter wider than snout length Habitat: Pelagic in lakes, slow or still waters Similar Species: Rainbow Smelt (Osmerus mordax), 75-80 lateral line scales Brook Silverside (Labidesthes sicculus) Elongated anal fin Images are not to scale 2 3 Centrarchidae Centrarchidae Redear Sunfish Distinguishing Features: (Lepomis microlophus) Redear Sunfish (Non-native) — — Sunfishes (Lepomis microlophus) Sunfishes Red on opercular flap No iridescent lines on cheek Long, pointed pectoral fins Bluegill (Native) Dark blotch at base (Lepomis macrochirus) of dorsal fin No red on opercular flap Coloring: Brownish-green to gray Blue-purple iridescence on cheek Bright red outer margin on opercular flap -

THE HYDROBIOTOPIC DIVERSITY of the LAKES of the LOWER PRUT RIVER, REPUBLIC of MOLDOVA Mihaela MUNTEANU (PILA)1, Silvius STANCIU
THE HYDROBIOTOPIC DIVERSITY OF THE LAKES OF THE LOWER PRUT RIVER, REPUBLIC OF MOLDOVA Mihaela MUNTEANU (PILA)1, Silvius STANCIU2 1PhD Student, „Dunărea de Jos” University of Galați, Romania, email: [email protected]. 2 PhD Professor „Dunărea de Jos”University of Galați, Romania, email: [email protected] Abstract This paper proposes the presentation and analysis of the fish fauna and the specific benthic resources of the Prut River and its tributaries: Costeşti-Stânca, Beleu, Manta. An important objective of the paper is to present the differences in hydrobiotop diversity in different riparian sectors. A healthy ecosystem has a rich ichthyofauna due to the diversity of vegetation and fish species capable of withstanding resistance to aggressive external factors and threats. The aquatic biodiversity of the Prut River is a major complex consisting of phytoplankton and zooplankton. To optimize production of fish, an important factor is maintaining the balance quantity and quality of the entire water system. From this point of view, the paper presented also quantitative aspects of zoobenthos, which is different in the analyzed areas, being influenced by the degree of pollution, the hydrological regime or the physico-chemical conditions. Research has revealed significant differences between different areas of the aquatic fauna of the Prut, mainly due to the human factor. The study is a preliminary one, preparing a broader analysis of the impact of the human factor in the aquatic areas of the Republic of Moldova. Identifying and analyzing the factors that have led to the modification of aquatic structure and diversity can serve as arguments for achieving viable measures for the protection and sustainable use of natural aquatic resources at national level. -

Identification of Pine Bunting T
Identification of Pine Bunting Daniele Occhiato he nominate subspecies of Pine Bunting tion between the two species in areas of sym- T Emberiza leucocephalos leucocephalos breeds patry on their Siberian breeding grounds. First- in a large part of Siberia from the western slopes generation hybrids, especially males, are gene- of the Ural (55° E) east to the Pacific, including rally distinctive and do not lead to confusion. Sakhalin and the Kuril Islands (c 155° E). It However, such hybrids are fertile, and back- ranges north to the Arctic Circle (66° N) and crosses with members of one or the other spe- south to northern Mongolia (50° N); a disjunct cies, or with other hybrids, lead to individuals in population breeds further south in the Altai, which evidence of hybridization is even more Tarbagatay, Ala Tau and Tien Shan mountain diluted, and often very difficult to detect in the ranges (45° N). A geographically isolated and field. In some cases, only careful in-hand exami- apparently sedentary subspecies, E l fronto, nation can reveal such hybrid characters. For breeds in northern Qinghai and Gansu prov- example, a study by Eugeny Panov (in Bradshaw inces, China (Cramp & Perrins 1994, Byers et al & Gray 1993) revealed that out of 239 adult 1995). The migratory nominate subspecies win- male Pine Bunting x Yellowhammer hybrids ters mostly in Afghanistan, Pakistan, north-west- studied in the hand in western Siberia, as many ern India, Nepal and northern China; less impor- as 58 were only identifiable as such by the yel- tant wintering areas include northern Iran, the low lesser underwing-coverts. -

Beyond Fish Edna Metabarcoding: Field Replicates Disproportionately Improve the Detection of Stream Associated Vertebrate Species
Metabarcoding and Metagenomics 5: 59–71 DOI 10.3897/mbmg.5.66557 Research Article Beyond fish eDNA metabarcoding: Field replicates disproportionately improve the detection of stream associated vertebrate species Till-Hendrik Macher1, Robin Schütz1, Jens Arle2, Arne J. Beermann1,3, Jan Koschorreck2, Florian Leese1,3 1 University of Duisburg-Essen, Aquatic Ecosystem Research, Universitätsstr. 5, 45141 Essen, Germany 2 German Environment Agency, Wörlitzer Platz 1, 06844 Dessau-Roßlau, Germany 3 University of Duisburg-Essen, Centre for Water and Environmental Research (ZWU), Universitätsstr. 3, 45141 Essen, Germany Corresponding author: Till-Hendrik Macher ([email protected]) Academic editor: Pieter Boets | Received 26 March 2021 | Accepted 10 June 2021 | Published 13 July 2021 Abstract Fast, reliable, and comprehensive biodiversity monitoring data are needed for environmental decision making and management. Recent work on fish environmental DNA (eDNA) metabarcoding shows that aquatic diversity can be captured fast, reliably, and non-invasively at moderate costs. Because water in a catchment flows to the lowest point in the landscape, often a stream, it can col- lect traces of terrestrial species via surface or subsurface runoff along its way or when specimens come into direct contact with water (e.g., when drinking). Thus, fish eDNA metabarcoding data can provide information on fish but also on other vertebrate species that live in riparian habitats. This additional data may offer a much more comprehensive approach for assessing vertebrate diversity at no additional costs. Studies on how the sampling strategy affects species detection especially of stream-associated communities, however, are scarce. We therefore performed an analysis on the effects of biological replication on both fish as well as (semi-)terrestrial species detection. -

Identification of Priority Areas for the Conservation of Stream Fish Assemblages: Implications for River Management in France A
Identification of Priority Areas for the Conservation of Stream Fish Assemblages: Implications for River Management in France A. Maire, P. Laffaille, J.F. Maire, L. Buisson To cite this version: A. Maire, P. Laffaille, J.F. Maire, L. Buisson. Identification of Priority Areas for the Conservation of Stream Fish Assemblages: Implications for River Management in France. River Research and Applications, Wiley, 2016, 33 (4), pp.524-537. 10.1002/rra.3107. hal-01426354 HAL Id: hal-01426354 https://hal.archives-ouvertes.fr/hal-01426354 Submitted on 2 Jul 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License IDENTIFICATION OF PRIORITY AREAS FOR THE CONSERVATION OF STREAM FISH ASSEMBLAGES: IMPLICATIONS FOR RIVER MANAGEMENT IN FRANCE A. MAIREa*,†, P. LAFFAILLEb,c, J.-F. MAIREd AND L. BUISSONb,e a Irstea; UR HYAX, Pôle Onema-Irstea Hydroécologie des plans d’eau; Centre d’Aix-en-Provence, Aix-en-Provence, France b CNRS; UMR 5245 EcoLab, (Laboratoire Ecologie Fonctionnelle et Environnement), Toulouse, France c Université de Toulouse, INP, UPS; EcoLab; ENSAT, Castanet Tolosan, France d ONERA, The French Aerospace Lab Composites Department, Châtillon, France e Université de Toulouse, INP, UPS; EcoLab, Toulouse, France ABSTRACT Financial and human resources allocated to biodiversity conservation are often limited, making it impossible to protect all natural places, and priority areas for protection must be identified. -
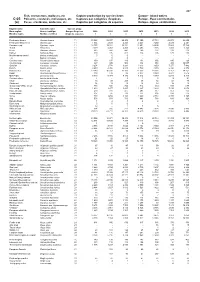
Fish, Crustaceans, Molluscs, Etc Capture Production by Species Items Europe
467 Fish, crustaceans, molluscs, etc Capture production by species items Europe - Inland waters C-05 Poissons, crustacés, mollusques, etc Captures par catégories d'espèces Europe - Eaux continentales (a) Peces, crustáceos, moluscos, etc Capturas por categorías de especies Europa - Aguas continentales English name Scientific name Species group Nom anglais Nom scientifique Groupe d'espèces 2009 2010 2011 2012 2013 2014 2015 Nombre inglés Nombre científico Grupo de especies t t t t t t t Freshwater bream Abramis brama 11 33 823 35 937 29 408 31 390 27 734 28 978 28 655 Freshwater breams nei Abramis spp 11 1 934 2 029 2 223 2 243 3 250 2 439 2 241 Common carp Cyprinus carpio 11 12 755 13 381 13 351 13 951 14 695 17 883 17 729 Tench Tinca tinca 11 3 019 4 053 2 455 2 253 1 686 1 491 1 328 Bleak Alburnus alburnus 11 682 547 441 269 231 213 219 Barbel Barbus barbus 11 300 195 412 224 205 170 188 Mediterranean barbel Barbus meridionalis 11 0 0 0 - - - - ...A Barbus cyclolepis 11 ... 0 0 - - - - Common nase Chondrostoma nasus 11 159 137 189 134 156 195 123 Crucian carp Carassius carassius 11 367 345 506 332 351 305 19 057 Goldfish Carassius auratus 11 3 254 2 778 3 293 3 613 3 653 7 277 7 482 Roach Rutilus rutilus 11 4 259 4 956 4 915 3 789 3 670 6 328 6 378 Roaches nei Rutilus spp 11 13 243 16 331 17 160 17 553 18 361 17 757 14 151 Rudd Scardinius erythrophthalmus 11 108 139 96 8 077 9 380 8 977 7 516 Orfe(=Ide) Leuciscus idus 11 5 040 5 374 5 330 4 812 5 979 6 240 5 316 Common dace Leuciscus leuciscus 11 4 .. -

Phylogeny and Classification of the Old World Emberizini
Available online at www.sciencedirect.com Molecular Phylogenetics and Evolution 47 (2008) 960–973 www.elsevier.com/locate/ympev Phylogeny and classification of the Old World Emberizini (Aves, Passeriformes) Per Alstro¨m a,b,*,1, Urban Olsson c,1, Fumin Lei d, Hai-tao Wang d,e, Wei Gao e, Per Sundberg c a Department of Vertebrate Zoology and Molecular Systematics Laboratory, Swedish Museum of Natural History, P.O. Box 50007, SE-104 05 Stockholm, Sweden b Department of Systematic Zoology, Evolutionary Biology Centre, Uppsala University, Norbyva¨gen 18 D, SE-752 36 Uppsala, Sweden c Department of Zoology, Go¨teborg University, Box 463, SE-405 30 Go¨teborg, Sweden d Institute of Zoology, Chinese Academy of Sciences, Datunlu B5, Changyang District, Beijing 100101, China e School of Life Sciences, Northeast Normal University, Changchun 130024, China Received 3 July 2007; revised 1 December 2007; accepted 11 December 2007 Available online 14 April 2008 Abstract The phylogeny of the avian genus Emberiza and the monotypic genera Latoucheornis, Melophus and Miliaria (collectively the Old World Emberizini), as well as representatives for the New World Emberizini, the circumpolar genera Calcarius and Plectrophenax and the four other generally recognized tribes in the subfamily Emberizinae was estimated based on the mitochondrial cytochrome b gene and introns 6–7 of the nuclear ornithine decarboxylase (ODC) gene. Our results support monophyly of the Old World Emberizini, but do not corroborate a sister relationship to the New World Emberizini. Calcarius and Plectrophenax form a clade separated from the other Emberizini. This agrees with previous studies, and we recommend the use of the name Calcariini. -

Observations of Asiatic Migrants in the Western Aleutians
JOURNAL OF FIELD ORNITHOLOGY Publishedby Association of Field Ornithologists VOL. 59, NO. 2 SPRING1988 P^GES101-208 j. Field Ornithol., 59(2):101-109 OBSERVATIONS OF ASIATIC MIGRANTS IN THE WESTERN ALEUTIANS MICHAEL H. TOVE 303 DunhagenPlace Cary, North Carolina27511 USA Abstract.--During the spring seasonsof 1977-1985, birders in the westernAleutians ob- servedlarge numbers of Asiaticmigrants whose occurrence was relatively predictable. These birdsregularly arrived during fair weather,sometimes in largemixed flocks, came from the southeast,and were exhaustedupon arrival. This set of observationsis consistentwith an expectedpattern of normaltransoceanic migration. Because non-passerines seemed to arrive in the early morningand passetinesin the afternoonand evening,a localizeddeparture pointis suggested.A simplemodel of migrationmechanics suggests that departurewas from the south-central Kurile Islands. OBSERVACIONES DE EMIGRANTES ASIATICOS EN LAS ALEUTIANAS DEL OESTE Resumen.--Desde el 1977-1985 observadores de aves localizados en las Aleutianas del oeste han notado durante la primavera la migraci6n de un gran ndmero de aves asifiticas.E1 patr6nde movimientoes predecible; llegan las avesde formaregular cuando las condiciones climato16gicasson buenas.Estas vienen del suroestey comoconsecuencia del viaje transø ocefinico11egan exhaustas. Dado el casode que las avesque no soncanoras parecen llegar temprano en la mafiana, mientras que los Passeriformesdurante la tarde y la noche,se sugiereen el trabajo un punto particular de partida localizadoen -

European Red List of Birds
European Red List of Birds Compiled by BirdLife International Published by the European Commission. opinion whatsoever on the part of the European Commission or BirdLife International concerning the legal status of any country, Citation: Publications of the European Communities. Design and layout by: Imre Sebestyén jr. / UNITgraphics.com Printed by: Pannónia Nyomda Picture credits on cover page: Fratercula arctica to continue into the future. © Ondrej Pelánek All photographs used in this publication remain the property of the original copyright holder (see individual captions for details). Photographs should not be reproduced or used in other contexts without written permission from the copyright holder. Available from: to your questions about the European Union Freephone number (*): 00 800 6 7 8 9 10 11 (*) Certain mobile telephone operators do not allow access to 00 800 numbers or these calls may be billed Published by the European Commission. A great deal of additional information on the European Union is available on the Internet. It can be accessed through the Europa server (http://europa.eu). Cataloguing data can be found at the end of this publication. ISBN: 978-92-79-47450-7 DOI: 10.2779/975810 © European Union, 2015 Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior written permission of the copyright holder. Printed in Hungary. European Red List of Birds Consortium iii Table of contents Acknowledgements ...................................................................................................................................................1 Executive summary ...................................................................................................................................................5 1. -

AHNELT H. 2008. Bestimmungsschlüssel Für Die In
Ahnelt H. 2008 Bestimmungsschlüssel 1 BESTIMMUNGSSCHLÜSSEL FÜR DIE IN ÖSTERREICH VORKOMMENDEN FISCHE HARALD AHNELT Department für Theoretische Biologie, Fakultät für Lebenswissenschaften, Universität Wien, Althanstrasse 14, 1090 Wien [email protected] Online: 10 September 2008 Zitiervorschlag: Ahnelt H. 2008 Bestimmungsschlüssel für die in Österreich vorkommenden Fische. http://homepage.univie.ac.at/harald.ahnelt/Harald_Ahnelts_Homepage/Publications.html [Download-Datum] Bestimmungsschlüssel heimischer Fische Dieser Bestimmungsschlüssel ist für die Fischarten Österreichs ausgelegt. Merkmale und Merkmalskombinationen können daher bei Anwendung auf Fische anderer Länder zu nicht korrekten Ergebnissen führen. Identification key for Austrian freshwater fishes This identification key should only be used for fishes from Austrian freshwaters. This key will possibly not work for fishes from other European countries. Nobody is perfect – schon gar nicht ein Bestimmungsschlüssel. Ein Bestimmungsschlüssel baut auf charakteristischen Merkmalen auf, er vereinfacht und kann keinen Anspruch auf Vollständigkeit erheben. Auch dieser Bestimmungsschlüssel ist nur ein Versuch ein komplexes System in einen übersichtliche Form zu bringen. Die Natur sieht aber oft anders aus. Die Bandbreite an Merkmalen ist bei vielen Arten groß. Manche Populationen sind an unterschiedliche Umweltbedingungen angepasst und bilden unterscheidbare ökologische Formen. Andere Populationen sind isoliert und einige davon sind systematisch noch ungenügend erforscht. Möglicherweise taucht ja in Österreich noch die eine oder andere neue Art auf. Sollte es einmal nicht passen, oder wenn sich ein Fehler eingeschlichen hat, ersuche ich um Information - [email protected] oder unter obiger Adresse. Verbesserungsvorschläge und Ergänzungen sind willkommen. Ahnelt H. 2008 Bestimmungsschlüssel 2 Einleitung 1858 erschien das Buch „Die Süßwasserfische der Österreichischen Monarchie mit Rücksicht auf die angrenzenden Länder“, verfasst von den Österreichern Johann Jakob Heckel und Rudolf Kner.