Identification of a Toxin–Antitoxin System That Contributes to Persister Formation by Reducing NAD in Pseudomonas Aeruginosa
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
FROM DNA to BEER Date Lesson Plan: Acquired and Passive Immunity Class Period
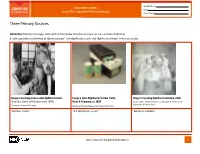
Student Name FROM DNA TO BEER Date Lesson Plan: Acquired and Passive Immunity Class Period Three Primary Sources Instruction: Examine the images and read their titles below. Based on what you can learn and infer from them: 1. Write your inferred definition of “diphtheria toxin,” “anti-diphtheritic serum,” and “diphtheria antitoxin” in the table below. Image 1. Injecting a horse with diphtheria toxin, Image 2. Anti-Diphtheritic Serum, Parke, Image 3. Injecting diphtheria antitoxin, 1895 New York City Health Department, 1940s Davis & Company, ca. 1898 Courtesy The Historical Medical Library of The College of Physicians of Philadelphia Courtesy Library of Congress Courtesy National Museum of American History “diphtheria toxin”: “anti-diphtheritic serum”: “diphtheria antitoxin”: http://www.nlm.nih.gov/fromdnatobeer 1 Student Name FROM DNA TO BEER Date Lesson Plan: Acquired and Passive Immunity Class Period Three Primary Sources 2. Describe or draw how the three images may be related. http://www.nlm.nih.gov/fromdnatobeer 2 FROM DNA TO BEER Lesson Plan: Acquired and Passive Immunity Teacher’s Three Primary Sources Instruction: Examine the images and read their titles below. Based on what you can learn and infer from them: 1. Write your inferred definition of “diphtheria toxin,” “anti-diphtheritic serum,” and “diphtheria antitoxin” in the table below. Image 1. Injecting a horse with diphtheria toxin, Image 2. Anti-Diphtheritic Serum, Parke, Image 3. Injecting diphtheria antitoxin, 1895 New York City Health Department, 1940s Davis & Company, -

Use of Ceragenins As a Potential Treatment for Urinary Tract Infections Urszula Wnorowska1, Ewelina Piktel1, Bonita Durnaś2, Krzysztof Fiedoruk3, Paul B
Wnorowska et al. BMC Infectious Diseases (2019) 19:369 https://doi.org/10.1186/s12879-019-3994-3 RESEARCH ARTICLE Open Access Use of ceragenins as a potential treatment for urinary tract infections Urszula Wnorowska1, Ewelina Piktel1, Bonita Durnaś2, Krzysztof Fiedoruk3, Paul B. Savage4 and Robert Bucki1* Abstract Background: Urinary tract infections (UTIs) are one of the most common bacterial infections. High recurrence rates and the increasing antibiotic resistance among uropathogens constitute a large social and economic problem in current public health. We assumed that combination of treatment that includes the administration ceragenins (CSAs), will reinforce the effect of antimicrobial LL-37 peptide continuously produced by urinary tract epithelial cells. Such treatment might be an innovative approach to enhance innate antibacterial activity against multidrug- resistant E. coli. Methods: Antibacterial activity measured using killing assays. Biofilm formation was assessed using crystal violet staining. Viability of bacteria and bladder epithelial cells subjected to incubation with tested agents was determined using MTT assays. We investigated the effects of chosen molecules, both alone and in combinations against four clinical strains of E. coli, obtained from patients diagnosed with recurrent UTI. Results: We observed that the LL-37 peptide, whose concentration increases at sites of urinary infection, exerts increased bactericidal effect against E. coli when combined with ceragenins CSA-13 and CSA-131. Conclusion: We suggest that the employment of combination of natural peptide LL-37 with synthetic analogs might be a potential solution to treat urinary tract infections caused by drug-resistant bacteria. Keywords: Urinary tract infection, LL-37 peptide, Ceragenins, Bacterial drug resistance Background trimethoprim/sulphamethoxazole, may no longer be used Urinary tract infections (UTIs) are one of the most com- for empiric treatment due to high resistance rates [7]. -

Antimicrobials: Killing Persisters While They Sleep
RESEARCH HIGHLIGHTS ANTIMICROBIALS Killing persisters while they sleep Bacterial persisters are a certain metabolites can enhance of mannitol enhanced gentamicin subpopulation of dormant cells the killing of both Gram-negative killing of persister cells by more than that have been implicated in a and Gram-positive persisters by two orders of magnitude. Similarly, it may be possible range of chronic and recurrent aminoglycosides. in mice that were implanted with to eradicate infections through their ability Aminoglycoside uptake into the catheters which had been colonized bacterial to survive antibiotic treatments. bacterium is energy dependent, by a uropathogenic E. coli strain, Although most cellular processes are leading the authors to investigate administration of mannitol together persisters in a completely shut down in persisters, whether metabolic stimulation with gentamicin reduced the viability clinical setting translation still occurs, albeit at a enhances the killing of persister of biofilm bacteria on the catheter by stimulating reduced rate, making the use of cells by increasing the uptake of by more than an order of magnitude. their underlying aminoglycoside antibiotics (which these antibiotics. They found that Finally, the authors tested whether metabolic activity target the ribosome) an attractive the addition of glucose, mannitol, metabolite addition enhanced option. However, aminoglycosides fructose or pyruvate increased the aminoglycoside killing of Gram- concurrently have only weak activity against this killing of isolated Escherichia coli positive bacteria; whereas mannitol, with antibiotic subpopulation of cells. Writing persisters by gentamicin, kanamycin glucose and pyruvate had no effect, treatment. in Nature, Collins and colleagues and streptomycin by more than three the addition of fructose enhanced now show that the addition of orders of magnitude. -

(ACIP) General Best Guidance for Immunization
Appendix 1: Glossary Adverse event. An undesirable medical condition that occurs following vaccination which might be truly caused by a vaccine, or it might be pure coincidence. Adverse reaction. An undesirable medical condition that has been demonstrated to be caused by a vaccine. Evidence for the causal relation is usually obtained through randomized clinical trials, controlled epidemiologic studies, isolation of the vaccine strain from the pathogenic site, or recurrence of the condition with repeated vaccination (i.e., rechallenge); synonyms include side effect and adverse effect. Adjuvant. A vaccine component distinct from the antigen that enhances the immune response to the antigen. Antitoxin. A solution of antibodies against a toxin. Antitoxin can be derived from either human (e.g., tetanus immune globulin) or animal (usually equine) sources (e.g., diphtheria and botulism antitoxin). Antitoxins are used to confer passive immunity and for treatment. Hyperimmune globulin (specific). Special preparations obtained from blood plasma from donor pools preselected for a high antibody content against a specific antigen (e.g., hepatitis B immune globulin, varicella-zoster immune globulin, rabies immune globulin, tetanus immune globulin, vaccinia immune globulin, cytomegalovirus immune globulin, botulism immune globulin). Immune globulin. A sterile solution containing antibodies, which are usually obtained from human blood. It is obtained by cold ethanol fractionation of large pools of blood plasma and contains 15%-18% protein. Intended for intramuscular administration, immune globulin is primarily indicated for routine maintenance of immunity among certain immunodeficient persons and for passive protection against measles and hepatitis A. General Best Practice Guidelines for Immunization: Appendix 1: Glossary 189 Immunobiologic. Antigenic substances (e.g., vaccines and toxoids) or antibody- containing preparations (e.g., globulins and antitoxins) from human or animal donors. -

Mechanisms of Staphylococcus Aureus Persistence And
MECHANISMS OF STAPHYLOCOCCUS AUREUS PERSISTENCE AND ERADICATION OF CHRONIC STAPHYLOCOCCAL INFECTIONS by Rebecca Yee A dissertation submitted to the Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland December, 2018 ABSTRACT Bacteria can exist in different phenotypic states depending on environmental conditions. Under stressed conditions, such as antibiotic exposure, bacteria can develop into persister cells that allow them to stay dormant until the stress is removed, when they can revert back to a growing state. The interconversion of non-growing persister cells and actively growing cells is the underlying basis of relapsing and chronic persistent infections. Eradication and better treatment of chronic, persistent infections caused by Staphylococcus aureus requires a multi- faceted approach, including a deeper understanding of how the bacteria persist under stressed conditions, regulate its cell death pathways, and development of novel drug therapies. Persisters were first discovered in the 1940s in a staphylococcal culture in which penicillin failed to kill a small subpopulation of the cells. Despite the discovery many decades ago, the specific mechanisms of Staphylococcus aureus persistence is largely unknown. Recently renewed interest has emerged due to the rise of chronic infections caused by pathogens such as M. tuberculosis, B. burgdorferi, S. aureus, P. aeruginosa, and E. coli. Treatments for chronic infections are lacking and antibiotic resistance is becoming a bigger issue. The goal of our research is to define the mechanisms involved in S. aureus persistence and cell death to improve our knowledge of genes and molecular pathways that can be used as targets for drugs to eradicate chronic infections. -

Diphtheria. In: Epidemiology and Prevention of Vaccine
Diphtheria Anna M. Acosta, MD; Pedro L. Moro, MD, MPH; Susan Hariri, PhD; and Tejpratap S.P. Tiwari, MD Diphtheria is an acute, bacterial disease caused by toxin- producing strains of Corynebacterium diphtheriae. The name Diphtheria of the disease is derived from the Greek diphthera, meaning ● Described by Hippocrates in ‘leather hide.’ The disease was described in the 5th century 5th century BCE BCE by Hippocrates, and epidemics were described in the ● Epidemics described in 6th century AD by Aetius. The bacterium was first observed 6th century in diphtheritic membranes by Edwin Klebs in 1883 and cultivated by Friedrich Löffler in 1884. Beginning in the early ● Bacterium first observed in 1900s, prophylaxis was attempted with combinations of toxin 1883 and cultivated in 1884 and antitoxin. Diphtheria toxoid was developed in the early ● Diphtheria toxoid developed 7 1920s but was not widely used until the early 1930s. It was in 1920s incorporated with tetanus toxoid and pertussis vaccine and became routinely used in the 1940s. Corynebacterium diphtheria Corynebacterium diphtheriae ● Aerobic gram-positive bacillus C. diphtheriae is an aerobic, gram-positive bacillus. ● Toxin production occurs Toxin production (toxigenicity) occurs only when the when bacillus is infected bacillus is itself infected (lysogenized) by specific viruses by corynebacteriophages (corynebacteriophages) carrying the genetic information for carrying tox gene the toxin (tox gene). Diphtheria toxin causes the local and systemic manifestations of diphtheria. ● Four biotypes: gravis, intermedius, mitis, and belfanti C. diphtheriae has four biotypes: gravis, intermedius, mitis, ● All isolates should be tested and belfanti. All biotypes can become toxigenic and cause for toxigenicity severe disease. -

Current Medical Literature American J. Digestive Diseases, Fort Wayne, Ind. Am. J. Roentgenol. & Rad. Therapy, Springfield
American Journal of Public Health, New York Current Medical Literature 33:925-1042 (Aug.) 1943 National Board of Health 1879-1883. W. G. Smillie.—p. 925. Preventive Medicine Program of United States Army. J. S. Simmons. AMERICAN —p. 931. Home Methods and Their Effect on Quality The Association to of Drying Palatability, Cooking library lends periodicals members the Association and Nutritive Value of Foods. Esther L. 941. and to Batchelder.—p. individual subscribers in continental United States and Canada Blood and Malaria Parasite Staining with Eosin Azure Méthylène Blue for a of be borrowed a time. period three days. Three journals may at Methods. R. D. 948. Periodicals are available from 1933 to date. for issues Lillie.—p. Requests of Radio Habits of Attend earlier date cannot be filled. should be Listening Mothers Who Well Baby Clinics. Requests accompanied by L. Murray and C. E. 952. cover if one and 18 if three Margaret Turner.—p. stamps to postage (6 cents cents periodicals Surveys of Nutrition of Populations: 2. Protein Nutrition of Rural are requested). Periodicals the American Medical Asso¬ published by Population in Middle Tennessee. J. B. E. W. ciation are not available for but can be on Youmans, Patton, lending supplied purchase W. R. Ruth Kern and Ruth 955. order. as a rule are the of can be Sutton, Steinkamp.—p. Reprints property authors and Field for Health Education Personnel. Minnie obtained for permanent possession only from them. Experience Krueger Oed. —p. 965. Titles marked with an asterisk (*) are abstracted below. Dehydration Procedures and Their Effect on Vitamin Retention. -

Eradication of Bacterial Persisters with Antibiotic-Generated Hydroxyl Radicals
Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals Sarah Schmidt Grant a,b,c,1, Benjamin B. Kaufmanna,c,d,1, Nikhilesh S. Chandd,e, Nathan Haseleya,d,f, and Deborah T. Hunga,b,c,d,2 aBroad Institute of MIT and Harvard, Cambridge, MA 02142; bDivision of Pulmonary and Critical Care Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA 02114; cDepartment of Molecular Biology and Center for Computational and Integrative Biology, Massachusetts General Hospital, Boston, MA 02114; dDepartment of Microbiology and Immunobiology, Harvard Medical School, Boston, MA 02115; eDepartment of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138; and fHarvard–MIT Division of Health Sciences and Technology, Cambridge, MA 02139 Edited by* Eric S. Lander, Broad Institute of MIT and Harvard, Cambridge, MA, and approved June 11, 2012 (received for review March 2, 2012) During Mycobacterium tuberculosis infection, a population of bac- terial cell numbers but do not sterilize the mouse (8). A plateau teria likely becomes refractory to antibiotic killing in the absence of is typically reached during which numbers of viable bacteria genotypic resistance, making treatment challenging. We describe stabilize. In addition to the mouse infection model, the inability an in vitro model capable of yielding a phenotypically antibi- to sterilize has been observed in the zebra fish (Mycobacterium otic-tolerant subpopulation of cells, often called persisters, within marinum), guinea pig (M. tuberculosis), and macrophage populations of Mycobacterium smegmatis and M. tuberculosis.We (M. tuberculosis) infection models (9–11). In vitro, the survival find that persisters are distinct from the larger antibiotic-suscepti- of a similar small subpopulation can also be observed when ble population, as a small drop in dissolved oxygen (DO) satura- a culture is exposed to high doses of antibiotics (12, 13). -

Humoral Immune Response Evaluation in Horses Vaccinated with Recombinant Clostridium Perfringens Toxoids Alpha and Beta for 12 Months
toxins Article Humoral Immune Response Evaluation in Horses Vaccinated with Recombinant Clostridium perfringens Toxoids Alpha and Beta for 12 Months Nayra F. Q. R. Freitas 1, Denis Y. Otaka 1, Cleideanny C. Galvão 1, Dayane M. de Almeida 1, Marcos R. A. Ferreira 2, Clóvis Moreira Júnior 2 , Marina M. M. H. Hidalgo 3, Fabricio R. Conceição 2 and Felipe M. Salvarani 1,* 1 Instituto de Medicina Veterinária, Universidade Federal do Pará, Castanhal CEP 68740-970, Brazil; [email protected] (N.F.Q.R.F.); [email protected] (D.Y.O.); [email protected] (C.C.G.); [email protected] (D.M.d.A.) 2 Centro de Desenvolvimento Tecnológico, Núcleo de Biotecnologia, Universidade Federal de Pelotas, Rio Grande do Sul CEP 96160-000, Brazil; [email protected] (M.R.A.F.); [email protected] (C.M.J.); [email protected] (F.R.C.) 3 Faculdade de Veterinária, Universidade Federal de Pelotas, Rio Grande do Sul CEP 96160-000, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-91-99-31-73-503 Abstract: In horses, Clostridium perfringens is associated with acute and fatal enterocolitis, which is caused by a beta toxin (CPB), and myonecrosis, which is caused by an alpha toxin (CPA). Although the most effective way to prevent these diseases is through vaccination, specific clostridial vaccines for horses against C. perfringens are not widely available. The aim of this study was to pioneer the immunization of horses with three different concentrations (100, 200 and 400 µg) of C. perfringens Citation: Freitas, N.F.Q.R.; Otaka, recombinant alpha (rCPA) and beta (rCPB) proteins, as well as to evaluate the humoral immune D.Y.; Galvão, C.C.; de Almeida, D.M.; response over 360 days. -

Phenol-Soluble Modulins Modulate Persister Cell Formation In
Phenol-Soluble Modulins Modulate Persister Cell Formation in Staphylococcus aureus Baldry, Mara; Bojer, Martin S; Najarzadeh, Zahra; Vestergaard, Martin; Meyer, Rikke Louise; Otzen, Daniel Erik; Ingmer, Hanne Published in: Frontiers in Microbiology DOI: 10.3389/fmicb.2020.573253 Publication date: 2020 Document version Publisher's PDF, also known as Version of record Document license: CC BY Citation for published version (APA): Baldry, M., Bojer, M. S., Najarzadeh, Z., Vestergaard, M., Meyer, R. L., Otzen, D. E., & Ingmer, H. (2020). Phenol-Soluble Modulins Modulate Persister Cell Formation in Staphylococcus aureus. Frontiers in Microbiology, 11, 573253. https://doi.org/10.3389/fmicb.2020.573253 Download date: 23. sep.. 2021 ORIGINAL RESEARCH published: 09 November 2020 doi: 10.3389/fmicb.2020.573253 Phenol-Soluble Modulins Modulate Persister Cell Formation in Staphylococcus aureus Mara Baldry 1†, Martin S. Bojer 1, Zahra Najarzadeh 2, Martin Vestergaard 1, Rikke Louise Meyer 2, Daniel Erik Otzen 2 and Hanne Ingmer 1* 1Department of Veterinary and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Frederiksberg, Denmark, 2Interdisciplinary Nanoscience Center (iNANO), Aarhus University, Aarhus, Denmark Edited by: Thomas Keith Wood, Pennsylvania State University (PSU), Staphylococcus aureus is a human pathogen that can cause chronic and recurrent United States infections and is recalcitrant to antibiotic chemotherapy. This trait is partly attributed to Reviewed by: Jie Feng, its ability to form persister cells, which are subpopulations of cells that are tolerant to lethal Lanzhou University Medical College, concentrations of antibiotics. Recently, we showed that the phenol-soluble modulins China (PSMs) expressed by S. aureus reduce persister cell formation. -

Persister Cells Form in the Plant Pathogen Xanthomonas Citri Subsp
microorganisms Article Persister Cells Form in the Plant Pathogen Xanthomonas citri subsp. citri under Different Stress Conditions Paula M. M. Martins 1,2 , Thomas K. Wood 1,* and Alessandra A. de Souza 2,* 1 Department of Chemical Engineering, Pennsylvania State University, University Park, PA 16802, USA; [email protected] 2 Biotechnology Laboratory, Centro de Citricultura Sylvio Moreira, Instituto Agronômico de Campinas, Rodovia Anhanguera Km 158, Cordeirópolis-SP 13490-000, Brazil * Correspondence: [email protected] (T.K.W.); [email protected] (A.A.d.S.) Abstract: Citrus canker disease, caused by the bacterium Xanthomonas citri subsp. citri is a constant threat to citrus-producing areas. Since it has no cure, agricultural practices to restrain its dissemination are essential to reduce the economic damage. Hence, increased knowledge of the basic aspects of X. citri biology could lead to more efficient management practices that can eliminate dormant bacteria in the field. The dormant cells, also referred to as persisters, are phenotypic variants with lowered metabolism, which in turn leads to tolerance to antimicrobials and undermines existing control approaches. We show here that X. citri forms persisters, identifying triggers for this phenotype, including antibiotics, high temperature, and metals (copper and zinc), which increase persistence rates by 10–100 times. The antioxidant N-acetylcysteine reduced copper and zinc-induced persisters, but not those induced by tetracycline, indicating that oxidative stress may be an important inducer of X. citri persistence. In addition, we found that metabolism-independent drugs like cisplatin and mitomycin C are able to eliminate X. citri persistent cells, as well as copper, at high concentrations. -

Controlling Bacterial Persister Cells and Biofilms by Synthetic Brominated Furanones
Syracuse University SURFACE Biomedical and Chemical Engineering - Dissertations College of Engineering and Computer Science 8-2013 Controlling Bacterial Persister Cells and Biofilms yb Synthetic Brominated Furanones Jiachuan Pan Follow this and additional works at: https://surface.syr.edu/bce_etd Part of the Chemical Engineering Commons Recommended Citation Pan, Jiachuan, "Controlling Bacterial Persister Cells and Biofilms yb Synthetic Brominated Furanones" (2013). Biomedical and Chemical Engineering - Dissertations. 68. https://surface.syr.edu/bce_etd/68 This Dissertation is brought to you for free and open access by the College of Engineering and Computer Science at SURFACE. It has been accepted for inclusion in Biomedical and Chemical Engineering - Dissertations by an authorized administrator of SURFACE. For more information, please contact [email protected]. ABSTRACT Bacteria are well known to obtain tolerance to antibiotics by forming multicellular structures, known as biofilms, and by entering dormancy and forming persister cells. Both mechanisms allow bacteria to tolerate antibiotics at concentrations hundreds to thousands of times higher than the lethal dose for regular planktonic cells of the same genotype. Persister formation increases in biofilms; thus, effective control of persister cells, especially those in biofilms is critically important to infection control. Over the past decades, a bacterial signaling system based on cell density, named quorum sensing (QS), has been found to regulate biofilm formation and, in Pseudomonas aeruginosa , the level of persistence. In this study, we characterized the effects of synthetic brominated furanones, a group of QS inhibitors, on the persistence of P. aeruginosa and Escherichia coli . Our results revealed that (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5 H)-one (BF8) can reduce persister formation in P.