Investigating the Potential for Environmental Perception and Adaptation in the Amoebaflagellate Naegleria Gruberi
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

UNIVERSITE DES ANTILLES Faculté Des Sciences Exactes Et Naturelles École Doctorale Pluridisciplinaire
UNIVERSITE DES ANTILLES Faculté des Sciences Exactes et Naturelles École Doctorale pluridisciplinaire Thèse de Doctorat En Physiologie-biologie des organismes-populations-interactions Mirna MOUSSA Les amibes libres pathogènes des eaux chaudes de la Guadeloupe, étude écologique, caractérisation moléculaire et prophylaxie des zones de baignade Sous la direction de : Jérôme GUERLOTTÉ, Professeur, Université des Antilles Antoine TALARMIN, Docteur, Institut Pasteur de la Guadeloupe Soutenue le 25 Septembre 2015 à l’Institut Pasteur de Guadeloupe N: Jury: GROS Olivier, Pr. Université des Antilles Président HÉCHARD Yann, Pr. Université de Poitiers Rapporteur YERA Hélène , MCU-PH Institut Cochin, Paris Rapporteur ALBINA Emmanuel, Dr. CIRAD Antilles Examinateur DESBOIS Nicole, Dr. CHU de Fort-de-France Examinateur GUERLOTTÉ Jérôme, Pr. Université des Antilles Examinateur 2 A ma famille et mes amis pour leur Amour et leur soutien au quotidien. Vous avez toujours su trouver les mots pour m’encourager dans les moments les plus difficiles. Je ne vous remercierai jamais assez pour tout ce que vous avez fait. Je pense à ma grand-mère en Syrie qui m’a appris la joie de vivre malgré les difficultés. 3 4 Remerciements Je souhaite tout d’abord exprimer ma profonde gratitude au Docteur Antoine Talarmin qui m’a permis de réaliser cette thèse au sein de l’Institut Pasteur de la Guadeloupe ainsi qu’au Professeur Jérôme Guerlotté qui m’a fait l’honneur d’être mon Directeur de thèse durant ces trois années. Je leur suis extrêmement reconnaissante de m’avoir fait confiance et de m’avoir donné des opportunités pour réussir dans ce domaine. Je les remercie pour leur patience, leurs conseils, leur soutien et leur disponibilité. -

Primary Amoebic Meningoencephalitis Amoebic Meningoencephalitis Is Primary Ś
PØEHLEDOVÉ PRÁCE PØEHLEDOVÉ JE NEGLERIÓZA VEREJNO-ZDRAVOTNÍCKYM PROBLÉMOM? IS PRIMARY AMOEBIC MENINGOENCEPHALITIS (NAEGLERIASIS) A PUBLIC HEALTH PROBLEM? KATARÍNA TRNKOVÁ, LUCIA MAĎAROVÁ, CYRIL KLEMENT Regionálny úrad verejného zdravotníctva so sídlom v Banskej Bystrici, odbor lekárskej mikrobiológie SOUHRN Neglerióza alebo primárna amébová meningoencefalitída (PAM) je zriedkavé ochorenie CNS, pôvodcom ktorého je vo¾ne žijúca meòavka Naegleria fowleri. Medzi stovkami vo¾ne žijúcich meòaviek sú známe i ïalšie rody, ktorých zástupcovia sú schopní infikovaś èloveka a vyvolaś u neho ochorenie. Za patogény sú považovaní zástupcovia rodov Acanthamoeba a Naegleria a druhy Balamuthia mandrillaris a Sappi- nia diploidea. Infekcie spôsobené týmito organizmami vyvolávajú u ¾udí syndrómy v rozsahu od akútnych fatálnych ochorení po chronické, tkanivá napadajúce infekcie s granulomatóznymi prejavmi. Epidemiológia, imunológia, patológia a klinické prejavy týchto infekcií sa vzájomne ve¾mi líšia. Príspevok podáva preh¾ad o pôvodcovi ochorenia PAM, o jeho morfológii, životnom cykle, ekológii ako aj o patogenéze, symptomatike a spôsoboch laboratórnej diagnostiky negleriózy. K¾úèové slová: neglerióza, primárna amébová meningoencefalitída, epidemiológia, laboratórna diagnostika Naegleria fowleri SUMMARY Naegleriasis or primary amoebic meningoencephalitis (PAM) is invariably an acute, often fulminant infection of CNS caused by Naegleria fowleri, a small, free-living amoeba. Pathogenic free-living amoebae can cause serious illnesses in humans. The amoe- HYGIENA bae belonging to the genus Naegleria, Acanthamoeba and Balamuthia mandrillaris and Sappinia diploidea produce syndromes in man ranging from acute fatal disease to chronic tissue invasion with granulomatous manifestation. The purpose of this report is to describe the clinical history, treatment, pathology and methods of laboratory diagnostic of naegleriasis. Key words: primary amoebic meningoencephalitis, naegleriasis, epidemiology, laboratory diagnostics of Naegleria fowleri ÈÍSLO 2 Úvod Obr. -

SNF Mobility Model: ICD-10 HCC Crosswalk, V. 3.0.1
The mapping below corresponds to NQF #2634 and NQF #2636. HCC # ICD-10 Code ICD-10 Code Category This is a filter ceThis is a filter cellThis is a filter cell 3 A0101 Typhoid meningitis 3 A0221 Salmonella meningitis 3 A066 Amebic brain abscess 3 A170 Tuberculous meningitis 3 A171 Meningeal tuberculoma 3 A1781 Tuberculoma of brain and spinal cord 3 A1782 Tuberculous meningoencephalitis 3 A1783 Tuberculous neuritis 3 A1789 Other tuberculosis of nervous system 3 A179 Tuberculosis of nervous system, unspecified 3 A203 Plague meningitis 3 A2781 Aseptic meningitis in leptospirosis 3 A3211 Listerial meningitis 3 A3212 Listerial meningoencephalitis 3 A34 Obstetrical tetanus 3 A35 Other tetanus 3 A390 Meningococcal meningitis 3 A3981 Meningococcal encephalitis 3 A4281 Actinomycotic meningitis 3 A4282 Actinomycotic encephalitis 3 A5040 Late congenital neurosyphilis, unspecified 3 A5041 Late congenital syphilitic meningitis 3 A5042 Late congenital syphilitic encephalitis 3 A5043 Late congenital syphilitic polyneuropathy 3 A5044 Late congenital syphilitic optic nerve atrophy 3 A5045 Juvenile general paresis 3 A5049 Other late congenital neurosyphilis 3 A5141 Secondary syphilitic meningitis 3 A5210 Symptomatic neurosyphilis, unspecified 3 A5211 Tabes dorsalis 3 A5212 Other cerebrospinal syphilis 3 A5213 Late syphilitic meningitis 3 A5214 Late syphilitic encephalitis 3 A5215 Late syphilitic neuropathy 3 A5216 Charcot's arthropathy (tabetic) 3 A5217 General paresis 3 A5219 Other symptomatic neurosyphilis 3 A522 Asymptomatic neurosyphilis 3 A523 Neurosyphilis, -

Mitochondrial DNA
GBE Missing Genes, Multiple ORFs, and C-to-U Type RNA Editing in Acrasis kona (Heterolobosea, Excavata) Mitochondrial DNA Cheng-Jie Fu1,*, Sanea Sheikh1, Wei Miao2, Siv G.E. Andersson3, and Sandra L. Baldauf1,* 1Program in Systematic Biology, Department of Organismal Biology, Evolutionary Biology Centre, Uppsala University, Sweden 2Key Laboratory of Aquatic Biodiversity and Conservation, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China 3Department of Molecular Evolution, Cell and Molecular Biology, Science for Life Laboratory, Biomedical Centre, Uppsala University, Sweden *Corresponding author: E-mail: [email protected], [email protected]; [email protected]. Accepted: August 18, 2014 Data deposition: The Acrasis kona mitochondrial genome sequence and annotation have been deposited at GenBank under the accession KJ679272. Downloaded from Abstract Discoba(Excavata)isanancientgroupof eukaryotes withgreatmorphological andecologicaldiversity.Unlikethe other major divisions http://gbe.oxfordjournals.org/ of Discoba (Jakobida and Euglenozoa), little is known about the mitochondrial DNAs (mtDNAs) of Heterolobosea. We have assembled a complete mtDNA genome from the aggregating heterolobosean amoeba, Acrasis kona, which consists of a single circular highly AT- rich (83.3%) molecule of 51.5 kb. Unexpectedly, A. kona mtDNA is missing roughly 40% of the protein-coding genes and nearly half of the transfer RNAs found in the only other sequenced heterolobosean mtDNAs, those of Naegleria spp. Instead, over a quarter of A. kona mtDNA consists of novel open reading frames. Eleven of the 16 protein-coding genes missing from A. kona mtDNA were identified in its nuclear DNA and polyA RNA, and phylogenetic analyses indicate that at least 10 of these 11 putative nuclear-encoded mitochondrial (NcMt) proteins arose by direct transfer from the mitochondrion. -

Issues of the Presence of Parasitic Protozoa in Surface Waters
E3S Web of Conferences 30, 01010 (2018) https://doi.org/10.1051/e3sconf/20183001010 Water, Wastewater and Energy in Smart Cities Issues of the presence of parasitic protozoa in surface waters Eliza Hawrylik1* 1 Department of Chemistry, Biology and Biotechnology, Faculty of Civil and Environmental Engineering, Bialystok University of Technology, Wiejska 45E Street, 15-351 Bialystok, Poland Abstract. Parasitic protozoa are very numerous organisms in the environment that play an important role in the spread of water-borne diseases. Water-borne epidemics caused by parasitic protozoa are noted throughout the world. Within these organisms, intestinal protozoa of the genera Cryptosporidium and Giardia are ones of the most serious health hazards for humans. This paper focuses on the problem of the presence of parasitic protozoa in surface waters. Characteristics of the most frequently recognized pathogens responsible for water-borne outbreaks were described, as well as sources of contamination and surface waters contamination due to protozoa of the genus Cryptosporidium and Giardia were presented. The methods of destroying the cysts and oocysts of parasitic protozoa used nowadays in the world were also presented in a review. 1 Occurrence of parasitic protozoa in surface water Parasitic protozoa are unicellular animal organisms that are very numerously isolated from natural waters, soils, food and media contaminated with animal manure and diseased humans. They are cosmopolitans, and are found in all countries of the world. They play an important role in the spread of water-borne diseases [1,2]. Water is the most common source of infection with protozoa, such as Entamoeba histolytica, Giardia intestinalis, Cryptosporidium sp., Cyclospora cayetanensis, Toxoplasma gondii. -

Toothless Aquatic Code Allows Deadly, Brain-Eating Zombie Amoeba to Flourish in Arizona Splash Pads and Water Playgrounds Sarah Pook*
IF WE ONLY HAD A BRAIN: Toothless Aquatic Code Allows Deadly, Brain-Eating Zombie Amoeba To Flourish in Arizona Splash Pads and Water Playgrounds Sarah Pook* I. INTRODUCTION It starts with a fever. A splitting headache. Vomiting, fatigue, an earache— then the secondary symptoms begin. Vision loss. Stiff neck. Lethargy, confusion, inability to walk, an aversion to light. Hallucinations. Doctors scramble to make a diagnosis, attempting treatment for bacterial meningitis, viral encephalitis, herpes, or other rare diseases, but nothing works.1 Finally, a coma. Death follows within three days.2 Diagnosis is usually done post-mortem: the culprit is primary amebic meningoencephalitis, or PAM.3 The disease is identified via a cerebrospinal fluid tap under a microscope. Peering in, you can see free living amoeba swimming around in the spinal fluid.4 * J.D. Candidate, 2020, Sandra Day O’Connor College of Law, Arizona State University; Editor-in-Chief, Arizona State Law Journal. I would like to thank Professor Tamara Herrera at Arizona State University for her insight and never-ending support as my faculty advisor, and Professor Marsha Howard at the University of Tulsa for her introduction to this fascinating topic. I would also like to thank my family for their love and encouragement. Finally, my heartfelt thanks to the wonderful Arizona State Law Journal staff and editors who bring our issues to publication—what a joy it is to work with all of you! 1. See Rikesh Baral & Binit Vaidya, Fatal Case of Amoebic Encephalitis Masquerading as Herpes, 5 OXFORD MED. CASE REPORTS. 146, 148 (2018), https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5934662/pdf/omy010.pdf [https://perma.cc/HSM6-WZZV]. -

Actions of Cannabinoids on Amoebae
Actions of Cannabinoids on Amoebae By Israa Al-hammadi This thesis is submitted for the degree of Doctor of Philosophy April 2020 Department of Biomedical and Life Sciences 2nd April 2020 To Whom It May Concern: Declaration Actions of cannabinoids on amoebae. This thesis has not been submitted in support of an application for another degree at this or any other university. It is the result of my own work and includes nothing that is the outcome of work done in collaboration except where specifically indicated. Many of the ideas in this thesis were the product of discussion with my supervisor Dr. Jackie Parry and Dr. Karen Wright. Israa Al-hammadi MSc. BSc. Table of contents Abstract 1 Chapter 1: General Introduction 3 1.1 Introduction 3 1.2 The Endocannabinoid System (ECS) in humans 3 1.2.1 General overview 3 1.2.2 Main ligands of the endocannabinoid system 3 1.2.2.1 Endocannabinoids 4 1.2.2.2 Phytocannabinoids 7 1.2.3 Main cannabinoid receptors 10 1.2.3.1. CB1 and CB2 receptors 10 1.2.3.2 The transient receptor potential vanilloid type 1 (TRPV1) 11 1.2.3.3 G-protein coupled receptor 55 (GPR55) 11 1.2.4 Other receptors 12 1.2.4.1 Peroxisome Proliferator-activated Receptors (PPARs) 12 1.2.4.2 Dopamine Receptors 14 1.2.4.3 Serotonin receptors (5-HT) 16 1.3. Existence of an endocannabinoid system in single-celled eukaryotes 17 1.3.1. Endocannabinoids in Tetrahymena 18 1.3.2. Enzymes in Tetrahymena 19 1.3.3. -

Taxonomy of Clinically Relevant Microorganisms*
OpenStax-CNX module: m58949 1 Taxonomy of Clinically Relevant Microorganisms* OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 4.0 1 Bacterial Pathogens The following tables list the species, and some higher groups, of pathogenic Eubacteria mentioned in the text. The classication of Bacteria, one of the three domains of life, is in constant ux as relationships become clearer through sampling of genetic sequences. Many groups at all taxonomic levels still have an undetermined relationship with other members of the phylogenetic tree of Bacteria. Bergey's Manual of Systematics of Archaea and Bacteria maintains a published list and descriptions of prokaryotic species. The tables here follow the taxonomic organization in the Bergey's Manual Taxonomic Outline.1 We have divided the species into tables corresponding to dierent bacterial phyla. The taxonomic rank of kingdom is not used in prokaryote taxonomy, so the phyla are the subgrouping below domain. Note that many bacterial phyla not represented by these tables. The species and genera are listed only under the class within each phylum. The names given to bacteria are regulated by the International Code of Nomenclature of Bacteria as maintained by the International Committee on Systematics or Prokaryotes. Phylum Actinobacteria Class Genus Species Related Diseases Corynebacterium diphtheriae Diphtheria Gardnerella vaginalis Bacterial vaginosis Micrococcus Opportunistic infections Actinobacteria Mycobacterium bovis Tuberculosis, primarily in cattle Mycobacterium leprae Hansen's disease continued on next page *Version 1.5: Apr 4, 2018 1:24 pm -0500 http://creativecommons.org/licenses/by/4.0/ 1Bergey's Manual Trust. Bergey's Manual of Systematics of Archaea and Bacteria, Taxonomic Outline. -

Is This an Outbreak?
Is This an Outbreak? A retrospective evaluation of syndromic surveillance for emerging infectious disease Is This an Outbreak? A retrospective evaluation of syndromic surveillance for emerging infectious disease Is dit een uitbraak? Een retrospectieve evaluatie van syndroomsurveillance voor nieuw opduikende infectieziekten Proefschrift ter verkrijging van de graad van doctor aan de Erasmus Universiteit Rotterdam op gezag van de rector magnificus Prof.dr. H.G. Schmidt en volgens besluit van het College voor Promoties. De openbare verdediging zal plaatsvinden op donderdag 9 december 2010 om 13.30 uur door © 2010 Kees van den Wijngaard, Zeist Cornelis Carolus van den Wijngaard ISBN 978 90 6464 426 9 Layout: GVO drukkers & vormgevers B.V. | Ponsen & Looijen geboren te Sittard Printed by: GVO drukkers & vormgevers B.V. | Ponsen & Looijen Reprints were made with permission of The American Public Health Association and Cambridge University Press. Publication of this thesis was financially supported by the National Institute for Public Health and the Environment (RIVM) and Erasmus University Rotterdam (EUR). Promotiecommissie Promotoren: Prof.dr. M.P.G. Koopmans Prof.dr. N.J.D. Nagelkerke Overige leden: Prof.dr. A. Osterhaus Prof.dr. J.H. Richardus Prof.dr. M.C.J.M. Sturkenboom Copromotor: Dr. W. van Pelt Contents Chapter 1 General Introduction 9 Chapter 2 Validation of Syndromic Surveillance for Respiratory Pathogen Activity Emerg Infect Dis 2008;14:917-25 19 Chapter 3 Detection of Excess Influenza Severity: Associating Respiratory Hospitalization -
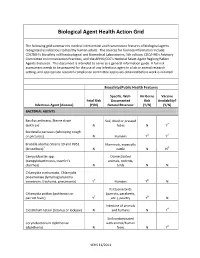
Biological Agent Health Action Grid
Biological Agent Health Action Grid The following grid summarizes medical intervention and transmission features of biological agents recognized as infectious to healthy human adults. The sources for foonote information include: CDC/NIH’s Biosafety in Microbiological and Biomedical Laboratories, 5th edition; CDC/HHS’s Advisory Committee on Immunization Practices; and the APHIS/CDC’s National Select Agent Registry/Select Agents Exclusion. This document is intended to serve as a general information guide. A full risk assessment needs to be prepared for the use of any infectious agent in a lab or animal research setting, and appropriate research compliance committee approvals obtained before work is initiated. Biosafety/Public Health Features Specific, Well- Air-Borne Vaccine Fetal Risk Documented Risk Availability? Infectious Agent (disease) (Y/N) Natural Reservoir (Y/N) (Y/N) BACTERIAL AGENTS Bacillus anthracis, Sterne strain Soil, dried or pressed (anthrax)1 N hides N Y2 Bordetella pertussis (whooping cough or pertussis) N Humans Y3 Y4 Brucella abortus Strains 19 and RB51 Mammals, especially (brucellosis)5 N cattle N N6 Campylobacter spp. Domesticated (campylobacteriosis, traveler's animals, rodents, diarrhea) N birds N N Chlamydia trachomatis, Chlamydia pneumoniae (lymphogranuloma venereum, trachoma, pneumonia) Y7 Humans Y8 N Psittacine birds Chlamydia psittaci (psittacosis or (parrots, parakeets, parrott fever) Y7 etc.), poultry Y8 N Intestine of animals Clostridium tetani (tetanus or lockjaw) N and humans N Y4 Soil contaminated Corynebacterium diphtheriae with animal/human (diphtheria) N feces N Y4 VEHS 11/2011 Specific, Well- Air-Borne Vaccine Fetal Risk Documented Risk Availability? Infectious Agent (disease) (Y/N) Natural Reservoir (Y/N) (Y/N) Francisella tularensis, subspecies Wild animals, esp. -

Downloaded from the Virulence Factor of Pathogenic Bacteria Database (VFDB)70
www.nature.com/scientificreports OPEN Investigation of potential pathogenicity of Willaertia magna by investigating the transfer of bacteria pathogenicity genes into its genome Issam Hasni1,2, Nisrine Chelkha1, Emeline Baptiste1, Mouh Rayane Mameri2, Joel Lachuer3,4,5, Fabrice Plasson2, Philippe Colson 1 & Bernard La Scola1* Willaertia magna c2c maky is a thermophilic amoeba closely related to the genus Naegleria. This free-living amoeba has the ability to eliminate Legionella pneumophila, which is an amoeba-resisting bacterium living in an aquatic environment. To prevent the proliferation of L. pneumophila in cooling towers, the use of W. magna as natural biocide has been proposed. To provide a better understanding of the W. magna genome, whole-genome sequencing was performed through the study of virulence factors and lateral gene transfers. This amoeba harbors a genome of 36.5 megabases with 18,519 predicted genes. BLASTp analyses reported protein homology between 136 W. magna sequences and amoeba-resistant microorganisms. Horizontal gene transfers were observed based on the basis of the phylogenetic reconstruction hypothesis. We detected 15 homologs of N. fowleri genes related to virulence, although these latter were also found in the genome of N. gruberi, which is a non-pathogenic amoeba. Furthermore, the cytotoxicity test performed on human cells supports the hypothesis that the strain c2c maky is a non-pathogenic amoeba. This work explores the genomic repertory for the frst draft genome of genus Willaertia and provides genomic data for further comparative studies on virulence of related pathogenic amoeba, N. fowleri. Heterolobosea is a rich class of species of heterotrophs, almost all free-living, containing pathogenic (e.g. -

Phylogeny of Deep-Level Relationships Within Euglenozoa Based on Combined Small Subunit and Large Subunit Ribosomal DNA Sequence
PHYLOGENY OF DEEP-LEVEL RELATIONSHIPS WITHIN EUGLENOZOA BASED ON COMBINED SMALL SUBUNIT AND LARGE SUBUNIT RIBOSOMAL DNA SEQUENCES by BING MA (Under the Direction of Mark A. Farmer) ABSTRACT My master’s research addressed questions about the evolutionary histories of Euglenozoa, with special attention given to deep-level relationships among taxa. The Euglenozoa are a putative early-branching assemblage of flagellated Eukaryotes, comprised primarily by three subgroups: euglenids, kinetoplastids and diplonemids. The goals of my research were to 1) evaluate the phylogenetic potential of large subunit ribosomal DNA (LSU rDNA) gene sequences as a molecular marker; 2) construct a phylogeny for the Euglenozoa to address their deep-level relationships; 3) provide morphological data of the flagellate Petalomonas cantuscygni to infer its evolutionary position in Euglenozoa. A dataset based on LSU rDNA sequences combined with SSU rDNA from thirty-nine taxa representing every subgroup of Euglenozoa and outgroup species was used for testing relationships within the Euglenozoa. Our results indicate that a) LSU rDNA is a useful marker to infer phylogeny, b) euglenids and diplonemdis are more closely related to one another than either is to the kinetoplatids and c) Petalomonas cantuscygni is closely related to the diplonemids. INDEX WORDS: Euglenozoa, phylogenetic inference, molecular evolution, 28S rDNA, LSU rDNA, 18S rDNA, SSU rDNA, euglenids, diplonemids, kinetoplastids, conserved core regions, expansion segments, TOL PHYLOGENY OF DEEP-LEVEL RELATIONSHIPS WITHIN EUGLENOZOA BASED ON COMBINED SMALL SUBUNIT AND LARGE SUBUNIT RIBOSOMAL DNA SEQUENCES by BING MA B. Med., Zhengzhou University, P. R. China, 2002 A Thesis Submitted to the Graduate Faculty of The University of Georgia in Partial Fulfillment of the Requirements for the Degree MASTER OF SCIENCE ATHENS, GEORGIA 2005 © 2005 Bing Ma All Rights Reserved PHYLOGENY OF DEEP-LEVEL RELATIONSHIPS WITHIN EUGLENOZOA BASED ON COMBINED SMALL SUBUNIT AND LARGE SUBUNIT RIBOSOMAL DNA SEQUENCES by BING MA Major Professor: Mark A.