Electron Microscopy and Antigenic Studies of Uncharacterized Viruses. I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Risk Groups: Viruses (C) 1988, American Biological Safety Association
Rev.: 1.0 Risk Groups: Viruses (c) 1988, American Biological Safety Association BL RG RG RG RG RG LCDC-96 Belgium-97 ID Name Viral group Comments BMBL-93 CDC NIH rDNA-97 EU-96 Australia-95 HP AP (Canada) Annex VIII Flaviviridae/ Flavivirus (Grp 2 Absettarov, TBE 4 4 4 implied 3 3 4 + B Arbovirus) Acute haemorrhagic taxonomy 2, Enterovirus 3 conjunctivitis virus Picornaviridae 2 + different 70 (AHC) Adenovirus 4 Adenoviridae 2 2 (incl animal) 2 2 + (human,all types) 5 Aino X-Arboviruses 6 Akabane X-Arboviruses 7 Alastrim Poxviridae Restricted 4 4, Foot-and- 8 Aphthovirus Picornaviridae 2 mouth disease + viruses 9 Araguari X-Arboviruses (feces of children 10 Astroviridae Astroviridae 2 2 + + and lambs) Avian leukosis virus 11 Viral vector/Animal retrovirus 1 3 (wild strain) + (ALV) 3, (Rous 12 Avian sarcoma virus Viral vector/Animal retrovirus 1 sarcoma virus, + RSV wild strain) 13 Baculovirus Viral vector/Animal virus 1 + Togaviridae/ Alphavirus (Grp 14 Barmah Forest 2 A Arbovirus) 15 Batama X-Arboviruses 16 Batken X-Arboviruses Togaviridae/ Alphavirus (Grp 17 Bebaru virus 2 2 2 2 + A Arbovirus) 18 Bhanja X-Arboviruses 19 Bimbo X-Arboviruses Blood-borne hepatitis 20 viruses not yet Unclassified viruses 2 implied 2 implied 3 (**)D 3 + identified 21 Bluetongue X-Arboviruses 22 Bobaya X-Arboviruses 23 Bobia X-Arboviruses Bovine 24 immunodeficiency Viral vector/Animal retrovirus 3 (wild strain) + virus (BIV) 3, Bovine Bovine leukemia 25 Viral vector/Animal retrovirus 1 lymphosarcoma + virus (BLV) virus wild strain Bovine papilloma Papovavirus/ -

The Poxviridae Comprise a Family of Complex Cytoplasmic Replicating
The Poxviridae comprise a family of complex cytoplasmic replicating DNA viruses that includes a number of important human and animal pathogens such as Variola virus, the causative agent of smallpox, Monkeypox virus, Cowpox virus and Vaccinia virus. Despite the successfully eradication of smallpox as a human disease by the worldwide vaccination campaigns 30 years ago, there nowadays still remains a considerable fear that Variola virus could be used as a bioweapon. Additionally, the re-emergence of zoonotic poxviruses such as the human Monkeypox virus outbreak that occurred in 2003 in the United States, Cowpox virus outbreaks in Europe and Vaccinia virus outbreaks in Brazil and India in the last years renewed the interest in the search for new anti- poxviral drugs. Besides, there is only one antiviral agent currently approved by FDA (Food and Drug Administration) for use against orthopoxviruses: cidofovir. However there are some concerns about its use, since cidofovir therapy can lead to renal and ocular toxicity, and also because cidofovir-resistant strains of camelpox, cowpox, monkeypox and vaccinia viruses have been isolated. In this context, the two poxvirus proteases, I7 and G1, arise as interesting protein targets for anti-poxvirus drugs development. In fact, it was previously showed that both I7 and G1 proteolysis activity are essential for viral replication and that these proteins have low identity with cellular proteins. These facts highlighted them as attractive antiviral targets. However, much essential knowledge of the molecular mechanisms of both proteases remains unexplored. This project aims to investigate more deeply both the structure and the function of these proteases. -
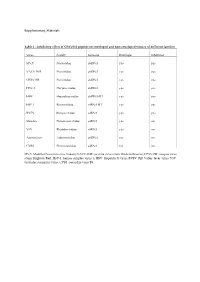
Inhibitory Effect of CPXV012 Peptide on Enveloped and Non-Enveloped Viruses of Different Families
Supplementary Materials Table 1 - Inhibitory effect of CPXV012 peptide on enveloped and non-enveloped viruses of different families Virus Family Genome Envelope Inhibition MVA Poxviridae dsDNA yes yes VACV-WR Poxviridae dsDNA yes yes CPXV-BR Poxviridae dsDNA yes yes HSV-1 Herpesviridae dsDNA yes yes HBV Hepadnaviridae dsDNA-RT yes yes HIV-1 Retroviridae ssRNA-RT yes yes RVFV Bunyaviridae ssRNA yes yes Measles Paramyxoviridae ssRNA yes no VSV Rhabdoviridae ssRNA yes no Adenovirus Adenoviridae dsDNA no no CVB3 Picornaviridae ssRNA no no MVA: Modified Vaccinia virus Ankara; VACV-WR: vaccinia virus strain Western Reserve; CPXV-BR: cowpox virus strain Brighton Red; HSV-1: herpes simplex virus-1; HBV: Hepatitis B virus; RVFV: Rift Valley fever virus; VSV: vesicular stomatitis virus; CVB3: coxsackie virus B3. A B 140 0.6 ) l 120 ro t 100 on 0.4 c g f 80 µ / o 60 % NR ( 0.2 1 - 40 T S 20 W 0 0.0 O 5 0 0 0 5 0 0 0 S 2 5 0 0 O 2 5 0 0 1 2 S 1 2 M M D D CPXV012 peptide [ g/mL] CPXV012 peptide [ g/mL] C ) . 30000 U . A ( e u 20000 l b r e t i t 10000 ll e C 0 0 50 100 150 200 CPXV012 peptide [ g/mL] Huh7.5 Vero Figure S1: CPXV012 peptide does not affect cell viability. (A-C) Cells were treated with the indicated concentrations of peptide or DMSO as vehicle control. S.E.M. of three independent experiments is shown. (A) Viability of MJS cells was measured by WST-1 assay or (B) Neutral Red uptake. -

Here, There, and Everywhere: the Wide Host Range and Geographic Distribution of Zoonotic Orthopoxviruses
viruses Review Here, There, and Everywhere: The Wide Host Range and Geographic Distribution of Zoonotic Orthopoxviruses Natalia Ingrid Oliveira Silva, Jaqueline Silva de Oliveira, Erna Geessien Kroon , Giliane de Souza Trindade and Betânia Paiva Drumond * Laboratório de Vírus, Departamento de Microbiologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais: Belo Horizonte, Minas Gerais 31270-901, Brazil; [email protected] (N.I.O.S.); [email protected] (J.S.d.O.); [email protected] (E.G.K.); [email protected] (G.d.S.T.) * Correspondence: [email protected] Abstract: The global emergence of zoonotic viruses, including poxviruses, poses one of the greatest threats to human and animal health. Forty years after the eradication of smallpox, emerging zoonotic orthopoxviruses, such as monkeypox, cowpox, and vaccinia viruses continue to infect humans as well as wild and domestic animals. Currently, the geographical distribution of poxviruses in a broad range of hosts worldwide raises concerns regarding the possibility of outbreaks or viral dissemination to new geographical regions. Here, we review the global host ranges and current epidemiological understanding of zoonotic orthopoxviruses while focusing on orthopoxviruses with epidemic potential, including monkeypox, cowpox, and vaccinia viruses. Keywords: Orthopoxvirus; Poxviridae; zoonosis; Monkeypox virus; Cowpox virus; Vaccinia virus; host range; wild and domestic animals; emergent viruses; outbreak Citation: Silva, N.I.O.; de Oliveira, J.S.; Kroon, E.G.; Trindade, G.d.S.; Drumond, B.P. Here, There, and Everywhere: The Wide Host Range 1. Poxvirus and Emerging Diseases and Geographic Distribution of Zoonotic diseases, defined as diseases or infections that are naturally transmissible Zoonotic Orthopoxviruses. Viruses from vertebrate animals to humans, represent a significant threat to global health [1]. -

Conventional and Unconventional Mechanisms for Capping Viral Mrna
REVIEWS Conventional and unconventional mechanisms for capping viral mRNA Etienne Decroly1, François Ferron1, Julien Lescar1,2 and Bruno Canard1 Abstract | In the eukaryotic cell, capping of mRNA 5′ ends is an essential structural modification that allows efficient mRNA translation, directs pre-mRNA splicing and mRNA export from the nucleus, limits mRNA degradation by cellular 5′–3′ exonucleases and allows recognition of foreign RNAs (including viral transcripts) as ‘non-self’. However, viruses have evolved mechanisms to protect their RNA 5′ ends with either a covalently attached peptide or a cap moiety (7‑methyl-Gppp, in which p is a phosphate group) that is indistinguishable from cellular mRNA cap structures. Viral RNA caps can be stolen from cellular mRNAs or synthesized using either a host- or virus-encoded capping apparatus, and these capping assemblies exhibit a wide diversity in organization, structure and mechanism. Here, we review the strategies used by viruses of eukaryotic cells to produce functional mRNA 5′-caps and escape innate immunity. Pre-mRNA splicing The cap structure found at the 5′ end of eukaryotic reactions involved in the viral RNA-capping process, A post-transcriptional mRNAs consists of a 7‑methylguanosine (m7G) moi‑ and the specific cellular factors that trigger a response modification of pre-mRNA, in ety linked to the first nucleotide of the transcript via a from the innate immune system. which introns are excised and 5′–5′ triphosphate bridge1 (FIG. 1a). The cap has several exons are joined in order to form a translationally important biological roles, such as protecting mRNA Capping, decapping and turnover of host RNA functional, mature mRNA. -

Why Do Poxviruses Still Matter? Zhilong Yang1,2* , Mark Gray2 and Lake Winter2
Yang et al. Cell Biosci (2021) 11:96 https://doi.org/10.1186/s13578-021-00610-8 Cell & Bioscience REVIEW Open Access Why do poxviruses still matter? Zhilong Yang1,2* , Mark Gray2 and Lake Winter2 Abstract Poxviruses comprise many members that infect both vertebrate and invertebrate animals, including humans. Despite the eradication of the historically notorious smallpox, poxviruses remain signifcant public health concerns and seri- ous endemic diseases. This short review briefy summarizes the present, historical, and future threats posed by pox- viruses to public health, wildlife and domestic animals, the role poxviruses have played in shaping modern medicine and biomedical sciences, the insight poxviruses have provided into complex life processes, and the utility of poxvi- ruses in biotechniques and in fghting other infectious diseases and cancers. It is anticipated that readers will appreci- ate the great merit and need for continued strong support of poxvirus research; research which benefts not only the expansion of fundamental biological knowledge but also the battle against diverse diseases. Keywords: Poxvirus, Vaccinia virus, Virology, Smallpox, Vaccine vector, Oncolytic therapy, Biodefense, Public health, Animal health Background of smallpox is unknown, it is believed that variola virus Te poxvirus family is a large family of double-stranded evolved from a rodent poxvirus [4]. While the lesions DNA viruses designated Poxviridae. Based on the Inter- found on the mummy of Egyptian Pharaoh Ramesses V national Committee on Taxonomy of Viruses (ICTV) (died 1145 BC) suggest that smallpox existed over Taxonomy 2019 release [1], Te Poxviridae comprises 3000 years ago [5, 6], the earliest written documenta- two subfamilies: Chordopoxvirinae (18 genera, 52 spe- tion of smallpox-like symptoms were from Ge Hong’s cies) and Entomopoxvarinae (4 genera, 30 species). -

Swinepox Virus
SWINEPOX VIRUS Prepared for the Swine Health Information Center By the Center for Food Security and Public Health, College of Veterinary Medicine, Iowa State University August 2015 SUMMARY Etiology • Swinepox virus (SwPV) is an enveloped DNA virus in the family Poxviridae; it is the only member of the genus Suipoxvirus and there is little genetic variability among strains. Cleaning and Disinfection • SwPV can persist in the environment, even in dry conditions. • SwPV is susceptible to most common forms of disinfectants including acid treatment, alcohols, aldehydes, alkalis, biguanides, halogens, oxidizing agents, phenolic compounds, and quaternary ammonium compounds. Epidemiology • Swine are the only natural hosts for SwPV. Humans are not affected; however, SwPV in pigs cannot be easily distinguished from vaccinia virus (VV), which is zoonotic. • SwPV is found worldwide. • Morbidity can be very high in pigs up to four months of age. Mortality is generally low (<5%), although congenital infections are frequently fatal. Transmission • SwPV is mechanically transmitted by the hog louse, Hematopinus suis, and possibly by biting flies (Stomoxys calcitrans) and black flies (Simuliidae). Pig-to-pig transmission can also occur. Infection in Swine/Pathogenesis • Classic pox disease is characterized by formation of macules, followed by progression to papules, vesicles, pustules, and crusts. Secondary bacterial infections can also occur. • Disease primarily occurs in pigs up to four months of age, while infection in adults is typically self-limiting. Diagnosis • SwPV may be cultivated in a range of host cells in vitro. Immunofluorescence and immunohistochemistry are used to detect SwPV antigens in infected epithelium. • A polymerase chain reaction (PCR) assay has been developed for rapid detection and differentiation from the clinically similar and zoonotic vaccinia virus (VV). -
Chapter 6 - Virology Viruses in Action!!
Chapter 6 - Virology Viruses in Action!! • Topics – Structure – Classification – Multiplication – Cultivation and replication – Nonviral infectious agent – Teratogenic/Oncogenic - Viruses have a host range. That is, viruses infect specific cells or tissues of specific hosts, or specific bacteria, Human or specific plants. Rhinovirus Common - Viral specificity refers to the specific Cold kinds of cells a virus can infect. It is regulated by the specificities of attachment, penetration and replication of the virus Properties of viruses A virion is an infectious virus particle - not all virus Viruses are not cells, do not have nuclei or particles are infectious mitochondria or ribosomes or other cellular Viruses are composed of a nucleic acid, RNA or DNA components. - never both . Viruses replicate or multiply. Viruses do not grow. All viruses have a protein coat or shell that surrounds Viruses replicate or multiply only within living cells. and protects the nucleic acid core. Viruses are obligate intracellular parasites . Some viruses have a lipid envelope or membrane surrounding a nucleocapsid core. The source of the The term virus was coined by Pasteur, and is from envelope is from the membranes of the host cell. the Latin word for poison. Some viruses package enzymes - e.g. RNA- Components of viruses - dependent-RNA polymerase or other enzymes - some do not package enzymes 1 Size comparison of viruses - how big are they? Structure • Size and morphology • Capsid • Envelope • Complex • Nucleic acid Mycoplasma? There are two major structures -

A Review of Virus Infections of Cetaceans and the Potential Impact of Morbilliviruses, Poxviruses and Papillomaviruses on Host Population Dynamics
DISEASES OF AQUATIC ORGANISMS Published October l l Dis Aquat Org 1 REVIEW A review of virus infections of cetaceans and the potential impact of morbilliviruses, poxviruses and papillomaviruses on host population dynamics Marie-Franqoise Van ~ressern'~~~',Koen Van waerebeekl, Juan Antonio Raga3 'Peruvian Centre for Cetacean Research (CEPEC), Jorge Chdvez 302, Pucusana, Lima 20. Peru 'Department of Vaccinology-Immunology, Faculty of Veterinary Medicine, University of Liege. Sart Tilman, 4000 Liege, Belgium 3Department of Animal Biology & Cavanilles Research Institute of Biodiversity and Evolutionary Biology, University of Valencia, Dr Moliner 50, 46100 Burjasot, Spain ABSTRACT. Viruses belonging to 9 farmlies have been detected in cetaceans. We critically review the clinical features, pathology and epidemiology of the diseases they cause. Cetacean morbillivirus (fam- ily Paramyxoviridae) induces a serious disease with a high mortality rate and persists in several popu- lation~.It may have long-term effects on the dynamics of cetacean populations either as enzoot~cinfec- tion or recurrent epizootics. The latter presumably have the more profound impact due to removal of sexually mature individuals. Members of the family Poxviridae infect several species of odontocetes, resulting in ring and tattoo skin lesions. Although poxviruses apparently do not induce a high mortality, circumstancial evidence suggests they may be lethal in young animals lacking protective immunity, and thus may negatively affect net recruitment. Papillomaviruses (family Papovaviridae) cause genital warts in at least 3 species of cetaceans. In 10% of male Burmeister's pal-poises Phocoena spinipinnis from Peru, lesions were sufficiently severe to at least hamper, if not impede, copulation. Members of the families Herpesviddae, Orthomyxoviridae and Rhabdov~ridaewere demonstrated in cetaceans suffer- ing serious illnesses, but with the exception of a 'porpoise herpesvirus' their causative role is still tentative. -

Viral Evolution: Accordion Adaptations Go Viral
RESEARCH HIGHLIGHTS Nature Reviews Microbiology | AOP, published online 10 September 2012; doi:10.1038/nrmicro2883 therefore viral replication. Vaccinia Although a higher gene copy virus, a Poxviridae member, has two number can be beneficial in the short Macmillan antagonists for PKR known as K3L term, in the long term the increased and E3L, which differ in their ability genome size would probably incur a to target PKR variants in different host substantial fitness cost. It is perhaps species. However, as PKR is antago- not a surprise, therefore, that in nized by a wide range of viruses, the viruses passaged ten times, 3–12% of protein undergoes rapid evolution, the sequenced K3L genes contained raising the question of how vaccinia a mutation that resulted in a single virus and its two PKR antagonists can amino acid substitution (His47Arg) keep up despite the low viral mutation which had been identified in a previ- rate. To investigate this problem, Elde ous genetic screen for K3L mutants et al. made use of the fact that K3L is with enhanced activity against a poor antagonist of human PKR; by human PKR. Importantly, in those propagating a strain of vaccinia virus viral genomes that had only a single lacking E3L (ΔE3L) in HeLa cells, they copy of K3L after ten passages, the could place a strong selective pressure frequency of the His47Arg mutation on K3L to adapt. The authors found had increased to 47%, suggesting that the ΔE3L virus initially replicated that there was a strong selective much more slowly than wild-type pressure to ensure that the His47Arg virus in HeLa cells over 48 hours, but allele was maintained in viruses that after six passages the rate of rep- with reduced K3L copy numbers. -

Informative Regions in Viral Genomes
viruses Article Informative Regions In Viral Genomes Jaime Leonardo Moreno-Gallego 1,2 and Alejandro Reyes 2,3,* 1 Department of Microbiome Science, Max Planck Institute for Developmental Biology, 72076 Tübingen, Germany; [email protected] 2 Max Planck Tandem Group in Computational Biology, Department of Biological Sciences, Universidad de los Andes, Bogotá 111711, Colombia 3 The Edison Family Center for Genome Sciences and Systems Biology, Washington University School of Medicine, Saint Louis, MO 63108, USA * Correspondence: [email protected] Abstract: Viruses, far from being just parasites affecting hosts’ fitness, are major players in any microbial ecosystem. In spite of their broad abundance, viruses, in particular bacteriophages, remain largely unknown since only about 20% of sequences obtained from viral community DNA surveys could be annotated by comparison with public databases. In order to shed some light into this genetic dark matter we expanded the search of orthologous groups as potential markers to viral taxonomy from bacteriophages and included eukaryotic viruses, establishing a set of 31,150 ViPhOGs (Eukaryotic Viruses and Phages Orthologous Groups). To do this, we examine the non-redundant viral diversity stored in public databases, predict proteins in genomes lacking such information, and used all annotated and predicted proteins to identify potential protein domains. The clustering of domains and unannotated regions into orthologous groups was done using cogSoft. Finally, we employed a random forest implementation to classify genomes into their taxonomy and found that the presence or absence of ViPhOGs is significantly associated with their taxonomy. Furthermore, we established a set of 1457 ViPhOGs that given their importance for the classification could be considered as markers or signatures for the different taxonomic groups defined by the ICTV at the Citation: Moreno-Gallego, J.L.; order, family, and genus levels. -

Viruses Infecting Reptiles
Viruses 2011, 3, 2087-2126; doi:10.3390/v3112087 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Viruses Infecting Reptiles Rachel E. Marschang Institut für Umwelt und Tierhygiene, University of Hohenheim, Garbenstr. 30, 70599 Stuttgart, Germany; E-Mail: [email protected]; Tel.: +49-711-459-22468; Fax: +49-711-459-22431 Received: 2 September 2011; in revised form: 19 October 2011 / Accepted: 21 October 2011 / Published: 1 November 2011 Abstract: A large number of viruses have been described in many different reptiles. These viruses include arboviruses that primarily infect mammals or birds as well as viruses that are specific for reptiles. Interest in arboviruses infecting reptiles has mainly focused on the role reptiles may play in the epidemiology of these viruses, especially over winter. Interest in reptile specific viruses has concentrated on both their importance for reptile medicine as well as virus taxonomy and evolution. The impact of many viral infections on reptile health is not known. Koch’s postulates have only been fulfilled for a limited number of reptilian viruses. As diagnostic testing becomes more sensitive, multiple infections with various viruses and other infectious agents are also being detected. In most cases the interactions between these different agents are not known. This review provides an update on viruses described in reptiles, the animal species in which they have been detected, and what is known about their taxonomic positions. Keywords: reptile; taxonomy; iridovirus; herpesvirus; adenovirus; paramyxovirus 1. Introduction Reptile virology is a relatively young field that has undergone rapid development over the past few decades.