Genetic Structure of Amblyomma Cajennense (Acari: Ixodidae) Populations Based on Mitochondrial Gene Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TICKS in RELATION to HUMAN DISEASES CAUSED by <I
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln U.S. Navy Research U.S. Department of Defense 1967 TICKS IN RELATION TO HUMAN DISEASES CAUSED BY RICKETTSIA SPECIES Harry Hoogstraal Follow this and additional works at: https://digitalcommons.unl.edu/usnavyresearch This Article is brought to you for free and open access by the U.S. Department of Defense at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in U.S. Navy Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. TICKS IN RELATION TO HUMAN DISEASES CAUSED BY RICKETTSIA SPECIES1,2 By HARRY HOOGSTRAAL Department oj Medical Zoology, United States Naval Medical Research Unit Number Three, Cairo, Egypt, U.A.R. Rickettsiae (185) are obligate intracellular parasites that multiply by binary fission in the cells of both vertebrate and invertebrate hosts. They are pleomorphic coccobacillary bodies with complex cell walls containing muramic acid, and internal structures composed of ribonucleic and deoxyri bonucleic acids. Rickettsiae show independent metabolic activity with amino acids and intermediate carbohydrates as substrates, and are very susceptible to tetracyclines as well as to other antibiotics. They may be considered as fastidious bacteria whose major unique character is their obligate intracellu lar life, although there is at least one exception to this. In appearance, they range from coccoid forms 0.3 J.I. in diameter to long chains of bacillary forms. They are thus intermediate in size between most bacteria and filterable viruses, and form the family Rickettsiaceae Pinkerton. They stain poorly by Gram's method but well by the procedures of Macchiavello, Gimenez, and Giemsa. -

Ehrlichiosis and Anaplasmosis Are Tick-Borne Diseases Caused by Obligate Anaplasmosis: Intracellular Bacteria in the Genera Ehrlichia and Anaplasma
Ehrlichiosis and Importance Ehrlichiosis and anaplasmosis are tick-borne diseases caused by obligate Anaplasmosis: intracellular bacteria in the genera Ehrlichia and Anaplasma. These organisms are widespread in nature; the reservoir hosts include numerous wild animals, as well as Zoonotic Species some domesticated species. For many years, Ehrlichia and Anaplasma species have been known to cause illness in pets and livestock. The consequences of exposure vary Canine Monocytic Ehrlichiosis, from asymptomatic infections to severe, potentially fatal illness. Some organisms Canine Hemorrhagic Fever, have also been recognized as human pathogens since the 1980s and 1990s. Tropical Canine Pancytopenia, Etiology Tracker Dog Disease, Ehrlichiosis and anaplasmosis are caused by members of the genera Ehrlichia Canine Tick Typhus, and Anaplasma, respectively. Both genera contain small, pleomorphic, Gram negative, Nairobi Bleeding Disorder, obligate intracellular organisms, and belong to the family Anaplasmataceae, order Canine Granulocytic Ehrlichiosis, Rickettsiales. They are classified as α-proteobacteria. A number of Ehrlichia and Canine Granulocytic Anaplasmosis, Anaplasma species affect animals. A limited number of these organisms have also Equine Granulocytic Ehrlichiosis, been identified in people. Equine Granulocytic Anaplasmosis, Recent changes in taxonomy can make the nomenclature of the Anaplasmataceae Tick-borne Fever, and their diseases somewhat confusing. At one time, ehrlichiosis was a group of Pasture Fever, diseases caused by organisms that mostly replicated in membrane-bound cytoplasmic Human Monocytic Ehrlichiosis, vacuoles of leukocytes, and belonged to the genus Ehrlichia, tribe Ehrlichieae and Human Granulocytic Anaplasmosis, family Rickettsiaceae. The names of the diseases were often based on the host Human Granulocytic Ehrlichiosis, species, together with type of leukocyte most often infected. -

An Insight Into the Ecobiology, Vector Significance and Control of Hyalomma Ticks (Acari: Ixodidae): a Review
Accepted Manuscript Title: AN INSIGHT INTO THE ECOBIOLOGY, VECTOR SIGNIFICANCE AND CONTROL OF HYALOMMA TICKS (ACARI: IXODIDAE): A REVIEW Authors: M.S. Sajid, A. Kausar, A. Iqbal, H. Abbas, Z. Iqbal, M.K. Jones PII: S0001-706X(18)30862-3 DOI: https://doi.org/10.1016/j.actatropica.2018.08.016 Reference: ACTROP 4752 To appear in: Acta Tropica Received date: 6-7-2018 Revised date: 10-8-2018 Accepted date: 12-8-2018 Please cite this article as: Sajid MS, Kausar A, Iqbal A, Abbas H, Iqbal Z, Jones MK, AN INSIGHT INTO THE ECOBIOLOGY, VECTOR SIGNIFICANCE AND CONTROL OF HYALOMMA TICKS (ACARI: IXODIDAE): A REVIEW, Acta Tropica (2018), https://doi.org/10.1016/j.actatropica.2018.08.016 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. AN INSIGHT INTO THE ECOBIOLOGY, VECTOR SIGNIFICANCE AND CONTROL OF HYALOMMA TICKS (ACARI: IXODIDAE): A REVIEW M. S. SAJID 1 2 *, A. KAUSAR 3, A. IQBAL 4, H. ABBAS 5, Z. IQBAL 1, M. K. JONES 6 1. Department of Parasitology, Faculty of Veterinary Science, University of Agriculture, Faisalabad-38040, Pakistan. 2. One Health Laboratory, Center for Advanced Studies in Agriculture and Food Security (CAS-AFS) University of Agriculture, Faisalabad-38040, Pakistan. -

Morphometrics of Amblyomma Mixtum in the State of Veracruz, Mexico
pathogens Article Morphometrics of Amblyomma mixtum in the State of Veracruz, Mexico Mariel Aguilar-Domínguez 1, Dora Romero-Salas 1,* , Sokani Sánchez-Montes 2, Ricardo Serna-Lagunes 3 , Greta Rosas-Saito 4, Anabel Cruz-Romero 1 and Adalberto A. Pérez de León 5,6 1 Laboratorio de Parasitología, rancho “Torreón del Molino”, Facultad de Medicina Veterinaria y Zootecnia, Universidad Veracruzana, Veracruz 91697, Mexico; [email protected] (M.A.-D.); [email protected] (A.C.-R.) 2 Facultad de Ciencias Biológicas y Agropecuarias Región Tuxpan, Universidad Veracruzana, Tuxpam 92870, Mexico; [email protected] 3 Laboratorio de Bioinformática y Bioestadística, Facultad de Ciencias Biológicas y Agropecuarias, Universidad Veracruzana, Córdoba 94945, Mexico; [email protected] 4 Red de Estudios Moleculares Avanzados, Instituto de Ecología, Xalapa 91073, Mexico; [email protected] 5 USDA-ARS Knipling-Bushland U.S. Veterinary Pest Genomics Center and Livestock Insects Research Laboratory, Kerrville, TX 78028, USA; [email protected] 6 USDA-ARS San Joaquin Valley Agricultural Sciences Center, Parlier, CA 93648, USA * Correspondence: [email protected]; Tel.: +52-(229)-9342075 Abstract: The tick Amblyomma mixtum is an ectoparasite of veterinary and public health importance because of its role as a vector of zoonotic pathogens. However, little is known about A. mixtum intraspecific variability and if morphological differentiation exists between populations across its geographic range. This study aimed to determine by electron microscopy the morphological vari- ability of A. mixtum populations in the state of Veracruz, which has a large livestock population Citation: Aguilar-Domínguez, M.; among states in Mexico. Forty male and 40 female A. mixtum collected from the 10 natural regions Romero-Salas, D.; Sánchez-Montes, S.; of Veracruz state were analyzed microscopically to accomplish main component analysis for each Serna-Lagunes, R.; Rosas-Saito, G.; sex. -
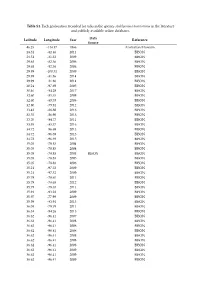
Table S1. Each Geolocation Recorded for Ticks in the Species Amblyomma Americanum in the Literature and Publicly Available Online Databases
Table S1. Each geolocation recorded for ticks in the species Amblyomma americanum in the literature and publicly available online databases. Data Latitude Longitude Year Reference Source 46.25 -114.17 1966 Australian Museum 28.31 -82.46 2011 BISON 28.51 -81.32 2009 BISON 29.68 -82.36 2006 BISON 29.68 -82.36 2006 BISON 29.99 -100.31 2009 BISON 29.99 -81.86 2014 BISON 29.99 -81.86 2014 BISON 30.24 -97.69 2005 BISON 30.46 -84.28 2017 BISON 32.60 -85.35 2008 BISON 32.60 -85.35 2008 BISON 32.80 -79.94 2012 BISON 33.42 -88.88 2016 BISON 33.55 -86.90 2013 BISON 33.70 -84.77 2011 BISON 33.95 -83.37 2016 BISON 34.72 -96.69 2013 BISON 34.72 -96.69 2013 BISON 34.72 -96.69 2015 BISON 35.05 -78.83 2004 BISON 35.05 -78.83 2004 BISON 35.05 -78.83 2004 BISON BISON 35.05 -78.83 2005 BISON 35.05 -78.83 2006 BISON 35.21 -97.32 2009 BISON 35.21 -97.32 2009 BISON 35.79 -78.65 2011 BISON 35.79 -78.65 2012 BISON 35.79 -78.65 2011 BISON 35.91 -93.22 2009 BISON 35.97 -77.99 2009 BISON 35.99 -83.94 2015 BISON 36.08 -79.79 2011 BISON 36.34 -94.26 2015 BISON 36.62 -96.41 2007 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2011 BISON 36.62 -96.41 2012 BISON 36.62 -96.41 2012 BISON 37.36 -77.05 2014 BISON 37.62 -84.87 2016 BISON 37.83 -78.28 2011 BISON 37.97 -85.70 2016 BISON 37.97 -85.70 2016 BISON 38.22 -75.31 2014 BISON 38.43 -88.43 -

Rocky Mountain Spotted Fever
Zoonosis Update Rocky Mountain spotted fever Ronald D. Warner, DVM, MPVM, PhD, DACVPM, and Wallace W. Marsh, MD, FAAP ocky Mountain spotted fever (RMSF), a classic SFG rickettsiae are transmitted by arthropods and cause Rmetazoonosis that involves both vertebrate and non- various illnesses worldwide, R rickettsii is the only one vertebrate reservoir hosts, is a seasonal disease of dogs known to be pathogenic for both animals and humans and humans in the Americas. The clinical illness was in the Western Hemisphere. first described among Native Americans, soldiers, and settlers in the Bitterroot River and Snake River valleys of Cycle of the Organism in Nature Montana and Idaho during the late 1890s, but remained and the Vectors unrecognized in dogs until the late 1970s. The causative Rickettsia rickettsii are maintained in nature by organism, Rickettsia rickettsii, was first described by transstadial passage within, and transovarial (vertical) Howard T. Ricketts in 1909 and is maintained in nature transmission between, generations of ixodid ticks. These by ixodid (hard-bodied) ticks via transmission to-and- ticks also vector R rickettsii to and from various rodent from various rodent reservoirs. As primary reservoir reservoirs and other small mammals. Naïve larval and hosts, the ticks vector R rickettsii to larger mammals; nymphal ticks become infected while feeding on small however, dogs and humans are the only ones that dis- rodents (eg, mice, voles, squirrels, or chipmunks) with play clinically recognizable illnesses. Rickettsia rickettsii acute rickettsemia.2,5 To enable transovarial transmission, are not naturally transmitted dog-to-dog, dog-to- female ticks need to ingest numerous rickettsiae or be human, or human-to-human. -

Analyzing the Potential Transmission of Tick-Borne Pathogens to Man
Revista da Sociedade Brasileira de Medicina Tropical ARTIGO 32(6):613-619, nov-dez, 1999. Report on ticks collected in the Southeast and Mid-West regions of Brazil: analyzing the potential transmission of tick-borne pathogens to man Relato sobre carrapatos coletados no Sudoeste e no Centro-oeste do Brasil analisando a potencial transmissão de microorganismos de carrapatos para o homem Luiz Tadeu Moraes Figueiredo1, Soraya Jabur Badra1, Luiz Eloy Pereira2 and Matias Pablo Juan Szabó3 Abstract Specimens of ticks were collected in 1993, 1996, 1997, and 1998, mostly from wild and domestic animals in the Southeast and Mid-West regions of Brazil. Nine species of Amblyommidae were identified: Anocentor nitens, Amblyomma cajennense, Amblyomma ovale, Amblyomma fulvum, Amblyomma striatum, Amblyomma rotundatum, Boophilus microplus, Boophilus annulatus, and Rhipicephalus sanguineus. The potential of these tick species as transmitters of pathogens to man was analyzed. A Flaviviridade Flavivirus was isolated from Amblyomma cajennense specimens collected from a sick capybara (Hydrochaeris hydrochaeris). Amblyomma cajennense is the main transmitter of Rickettsia rickettsii (=R. rickettsi), the causative agent of spotted fever in Brazil. Wild mammals, mainly capybaras and deer, infested by ticks and living in close contact with cattle, horses and dogs, offer the risk of transmission of wild zoonosis to these domestic animals and to man. Key-words: Brazilian ticks. Tick-borne pathogens. Resumo Foram coletados espécimes de carrapatos em 1993, 1996, 1997, e 1998, principalmente de animais selvagens e domésticos, nas Regiões Sudeste e Centro-oeste do Brasil. Nove espécies de Amblyommidae foram identificadas: Anocentor Nitens, Amblyomma cajennense, Amblyomma ovale, Amblyomma fulvum, Amblyomma striatum, Amblyomma rotundatum, Boophilus microplus, Boophilus annulatus e Rhipicephalus sanguineus. -

First Evidence of Ehrlichia Minasensis Infection in Horses from Brazil
pathogens Article First Evidence of Ehrlichia minasensis Infection in Horses from Brazil Lívia S. Muraro 1, Aneliza de O. Souza 2, Tamyres N. S. Leite 2, Stefhano L. Cândido 3, Andréia L. T. Melo 4, Hugo S. Toma 5 , Mariana B. Carvalho 4, Valéria Dutra 3, Luciano Nakazato 3, Alejandro Cabezas-Cruz 6 and Daniel M. de Aguiar 1,* 1 Laboratory of Virology and Rickettsial Infections, Veterinary Hospital, Federal University of Mato Grosso (UFMT), Av. Fernando Correa da Costa 2367, Cuiabá 78090-900, Brazil; [email protected] 2 Veterinary Clinical Laboratory, Department of Veterinary Clinics, University of Cuiabá (UNIC), Av. Manoel José de Arruda 3100, Cuiabá 78065-900, Brazil; [email protected] (A.d.O.S.); [email protected] (T.N.S.L.) 3 Laboratory of Microbiology and Molecular Biology, Veterinary Hospital of the Faculty of Veterinary Medicine, Federal University of Mato Grosso (UFMT), Av. Fernando Correa da Costa 2367, Cuiabá 78090-900, Brazil; [email protected] (S.L.C.); [email protected] (V.D.); [email protected] (L.N.) 4 Veterinary of Clinical, Veterinary Medicine College, University of Cuiabá (UNIC), Av. Manoel José de Arruda 3100, Cuiabá 78065-900, Brazil; [email protected] (A.L.T.M.); [email protected] (M.B.C.) 5 Veterinary Medicine Department, Federal University of Lavras (UFLA), Campus Universitário, Mailbox 3037, Lavras 37200-000, Brazil; hugo.toma@ufla.br 6 Anses, INRAE, Ecole Nationale Vétérinaire d’Alfort, UMR BIPAR, Laboratoire de Santé Animale, F-94700 Maisons-Alfort, France; [email protected] * Correspondence: [email protected] Citation: Muraro, L.S.; Souza, A.d.O.; Leite, T.N.S.; Cândido, S.L.; Melo, Abstract: The genus Ehrlichia includes tick-borne bacterial pathogens affecting humans, domestic and A.L.T.; Toma, H.S.; Carvalho, M.B.; wild mammals. -

Ticks and Tick-Borne Infections: Complex Ecology, Agents, and Host Interactions
veterinary sciences Review Ticks and Tick-Borne Infections: Complex Ecology, Agents, and Host Interactions Stephen K. Wikel Department of Medical Sciences, Frank H. Netter, M.D., School of Medicine, Quinnipiac University, Hamden, CT 06518, USA; [email protected]; Tel.: +1-203-626-9231 Received: 16 May 2018; Accepted: 15 June 2018; Published: 20 June 2018 Abstract: Ticks transmit the most diverse array of infectious agents of any arthropod vector. Both ticks and the microbes they transmit are recognized as significant threats to human and veterinary public health. This article examines the potential impacts of climate change on the distribution of ticks and the infections they transmit; the emergence of novel tick-borne pathogens, increasing geographic range and incidence of tick-borne infections; and advances in the characterization of tick saliva mediated modulation of host defenses and the implications of those interactions for transmission, establishment, and control of tick infestation and tick-borne infectious agents. Keywords: ticks; tick-borne diseases; emerging and resurging pathogens; zoonoses; vector ecology; climate change; tick saliva; tick-host-pathogen interactions; host immune defenses; immunomodulation 1. Introduction Zoonotic diseases are a significant negative impact upon global public health that is increasing. Zoonotic diseases are defined as infections shared in nature between humans and other vertebrate animal species [1], characterized by transmission between species of organisms infecting humans that are enzootic in other animal species [2]. Mutually transmissible infections between humans and other animal species are thought to have arisen with the development of agriculture and the resulting contact of more densely populated human communities with pathogens of domesticated and wild animal species [3]. -

09 Jose Brites.Indd
Veterinary World, EISSN: 2231-0916 REVIEW ARTICLE Available at www.veterinaryworld.org/Vol.8/March-2015/9.pdf Open Access Tick-borne infections in human and animal population worldwide José Brites-Neto1, Keila Maria Roncato Duarte2 and Thiago Fernandes Martins3 1. Department of Public Health, Americana, São Paulo, Brazil; 2. Department of Genetics and Animal Reproduction, Institute of Animal Science, Nova Odessa, São Paulo, Brazil; 3. Department of Preventive Veterinary Medicine, Faculty of Veterinary Medicine and Animal Sciences, University of São Paulo, São Paulo, Brazil. Corresponding author: José Brites-Neto, e-mail: [email protected], KMRD: [email protected], TFM: [email protected] Received: 14-11-2014, Revised: 20-01-2015, Accepted: 25-01-2015, Published online: 12-03-2015 doi: 10.14202/vetworld.2015.301-315. How to cite this article: Brites-Neto J, Duarte KMR, Martins TF (2015) Tick- borne infections in human and animal population worldwide, Veterinary World 8(3):301-315. Abstract The abundance and activity of ectoparasites and its hosts are affected by various abiotic factors, such as climate and other organisms (predators, pathogens and competitors) presenting thus multiples forms of association (obligate to facultative, permanent to intermittent and superficial to subcutaneous) developed during long co-evolving processes. Ticks are ectoparasites widespread globally and its eco epidemiology are closely related to the environmental conditions. They are obligatory hematophagous ectoparasites and responsible as vectors or reservoirs at the transmission of pathogenic fungi, protozoa, viruses, rickettsia and others bacteria during their feeding process on the hosts. Ticks constitute the second vector group that transmit the major number of pathogens to humans and play a role primary for animals in the process of diseases transmission. -

82179247.Pdf
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Experimental Parasitology 128 (2011) 324–327 Contents lists available at ScienceDirect Experimental Parasitology journal homepage: www.elsevier.com/locate/yexpr Failure of the Amblyomma cajennense nymph to become infected by Theileria equi after feeding on acute or chronically infected horses ⇑ Múcio F.B. Ribeiro , Júlia A.G. da Silveira, Camila V. Bastos Departamento de Parasitologia, ICB-UFMG, Belo Horizonte, Av. Antônio Carlos, 6627 Belo Horizonte, Minas Gerais, Brazil article info abstract Article history: Tick-borne diseases in horses are caused by the intraerythrocytic protozoan parasites Theileria equi Received 27 August 2010 and Babesia caballi. Although T. equi is highly endemic in Latin America, the New World vector of this Received in revised form 28 March 2011 important parasite is controversial. The aim of this study was to test the ability of nymph Amblyomma Accepted 29 March 2011 cajennense ticks acquire infection by T. equi following feeding on infected horses. Three experiments Available online 9 April 2011 were performed: tick acquisition of T. equi from an experimentally infected horse, tick acquisition of T. equi from naturally infected foals and tick acquisition of T. equi from a chronically infected horse. Keywords: A. cajennense adults were dissected and salivary glands were collected in aliquots. Methyl green pyro- Theileria equi nin staining of the salivary glands did not show the presence of hypertrophy of acini or cell nuclei Amblyomma cajennense Transmission normally suggestive of Theileria spp. infection. The pools of salivary glands were negative for Theileria DNA in nested PCR assays. -

Vector Borne Diseases Technical Bulletin
Vector-Borne Disease Tick-Borne Diseases of the United States Anaplasmosis Figure 1: Geographic map of Anaplasmosis reported to CDC, U.S., Figure 2: Morulae detected in a granulocyte on a peripheral 2016 (1). blood smear, associated with A. phagocytophilum infection. Photo/Bobbi S. Pritt, Mayo Clinic (2). Pathogen(s): Anaplasma phagocytophilum (formerly Human Granulocytic Ehrlichiosis, HGE) Location: Upper Midwest and Northeast United States overlapping with the geographic distribution of Lyme disease and other Blacklegged tick (Ixodes scapularis) transmitted diseases. Peak Infections: June through August Vector: Blacklegged ticks (Ixodes scapularis) Incubation Period: 5-14 days Signs & Symptoms: Fever, chills, rigors, severe headache, myalgia, gastrointestinal symptoms (nausea, vomiting, diarrhea, and anorexia) and rash (<10%). Few people will develop all symptom and the number and combination of symptoms varies greatly for each patient. Laboratory findings: Mild anemia, Leukopenia (characterized by relative and absolute lymphopenia and left shift), Thrombocytopenia, mild to moderate elevations in hepatic transaminases. PCR testing is most sensitive in during the first week of illness. Antibody based testing for rise in IgM (increase 2-3 days after illness) and IgG (typically up to 4-fold increase 7-10 days after illness). Samples should be taken within the first week of illness and a second sample should be taken 2-4 weeks later. MDL Test Code(s): 441 Ehrlichia chaffeensis (HME) & Anaplasma phgocytophilum (HGE) by Real-Time PCR 439 Anaplasma phagocytophilum IgG/IgM by IFA Treatment: Adults: 100 mg Doxycycline twice per day (100 mg/dose max), orally or IV for 10-14 days. Children (weighing <100 lbs, 45.4 kg): 2.2 mg/kg per dose Doxycycline (100 mg/dose max), twice per day, orally or IV for 10-14 days.