Exploring the Role of Endoplasmic Reticulum Stress in Hepatocellular Carcinoma Through Mining of the Human Protein Atlas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]](https://docslib.b-cdn.net/cover/8283/computational-genome-wide-identification-of-heat-shock-protein-genes-in-the-bovine-genome-version-1-peer-review-2-approved-1-approved-with-reservations-88283.webp)
Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]
F1000Research 2018, 7:1504 Last updated: 08 AUG 2021 RESEARCH ARTICLE Computational genome-wide identification of heat shock protein genes in the bovine genome [version 1; peer review: 2 approved, 1 approved with reservations] Oyeyemi O. Ajayi1,2, Sunday O. Peters3, Marcos De Donato2,4, Sunday O. Sowande5, Fidalis D.N. Mujibi6, Olanrewaju B. Morenikeji2,7, Bolaji N. Thomas 8, Matthew A. Adeleke 9, Ikhide G. Imumorin2,10,11 1Department of Animal Breeding and Genetics, Federal University of Agriculture, Abeokuta, Nigeria 2International Programs, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, 14853, USA 3Department of Animal Science, Berry College, Mount Berry, GA, 30149, USA 4Departamento Regional de Bioingenierias, Tecnologico de Monterrey, Escuela de Ingenieria y Ciencias, Queretaro, Mexico 5Department of Animal Production and Health, Federal University of Agriculture, Abeokuta, Nigeria 6Usomi Limited, Nairobi, Kenya 7Department of Animal Production and Health, Federal University of Technology, Akure, Nigeria 8Department of Biomedical Sciences, Rochester Institute of Technology, Rochester, NY, 14623, USA 9School of Life Sciences, University of KwaZulu-Natal, Durban, 4000, South Africa 10School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, 30032, USA 11African Institute of Bioscience Research and Training, Ibadan, Nigeria v1 First published: 20 Sep 2018, 7:1504 Open Peer Review https://doi.org/10.12688/f1000research.16058.1 Latest published: 20 Sep 2018, 7:1504 https://doi.org/10.12688/f1000research.16058.1 Reviewer Status Invited Reviewers Abstract Background: Heat shock proteins (HSPs) are molecular chaperones 1 2 3 known to bind and sequester client proteins under stress. Methods: To identify and better understand some of these proteins, version 1 we carried out a computational genome-wide survey of the bovine 20 Sep 2018 report report report genome. -

Chromosomal Aberrations in Head and Neck Squamous Cell Carcinomas in Norwegian and Sudanese Populations by Array Comparative Genomic Hybridization
825-843 12/9/08 15:31 Page 825 ONCOLOGY REPORTS 20: 825-843, 2008 825 Chromosomal aberrations in head and neck squamous cell carcinomas in Norwegian and Sudanese populations by array comparative genomic hybridization ERIC ROMAN1,2, LEONARDO A. MEZA-ZEPEDA3, STINE H. KRESSE3, OLA MYKLEBOST3,4, ENDRE N. VASSTRAND2 and SALAH O. IBRAHIM1,2 1Department of Biomedicine, Faculty of Medicine and Dentistry, University of Bergen, Jonas Lies vei 91; 2Department of Oral Sciences - Periodontology, Faculty of Medicine and Dentistry, University of Bergen, Årstadveien 17, 5009 Bergen; 3Department of Tumor Biology, Institute for Cancer Research, Rikshospitalet-Radiumhospitalet Medical Center, Montebello, 0310 Oslo; 4Department of Molecular Biosciences, University of Oslo, Blindernveien 31, 0371 Oslo, Norway Received January 30, 2008; Accepted April 29, 2008 DOI: 10.3892/or_00000080 Abstract. We used microarray-based comparative genomic logical parameters showed little correlation, suggesting an hybridization to explore genome-wide profiles of chromosomal occurrence of gains/losses regardless of ethnic differences and aberrations in 26 samples of head and neck cancers compared clinicopathological status between the patients from the two to their pair-wise normal controls. The samples were obtained countries. Our findings indicate the existence of common from Sudanese (n=11) and Norwegian (n=15) patients. The gene-specific amplifications/deletions in these tumors, findings were correlated with clinicopathological variables. regardless of the source of the samples or attributed We identified the amplification of 41 common chromosomal carcinogenic risk factors. regions (harboring 149 candidate genes) and the deletion of 22 (28 candidate genes). Predominant chromosomal alterations Introduction that were observed included high-level amplification at 1q21 (harboring the S100A gene family) and 11q22 (including Head and neck squamous cell carcinoma (HNSCC), including several MMP family members). -

Gene Targeting Therapies (Roy Alcalay)
Recent Developments in Gene - Targeted Therapies for Parkinson’s Disease Roy Alcalay, MD, MS Alfred and Minnie Bressler Associate Professor of Neurology Division of Movement Disorders Columbia University Medical Center Disclosures Funding: Dr. Alcalay is funded by the National Institutes of Health, the DOD, the Michael J. Fox Foundation and the Parkinson’s Foundation. Dr. Alcalay receives consultation fees from Genzyme/Sanofi, Restorbio, Janssen, and Roche. Gene Localizations Identified in PD Gene Symbol Protein Transmission Chromosome PARK1 SNCA α-synuclein AD 4q22.1 PARK2 PRKN parkin (ubiquitin ligase) AR 6q26 PARK3 ? ? AD 2p13 PARK4 SNCA triplication α-synuclein AD 4q22.1 PARK5 UCH-L1 ubiquitin C-terminal AD 4p13 hydrolase-L1 PARK6 PINK1 PTEN-induced kinase 1 AR 1p36.12 PARK7 DJ-1 DJ-1 AR 1p36.23 PARK8 LRRK2 leucine rich repeat kinase 2 AD 12q12 PARK9 ATP13A2 lysosomal ATPase AR 1p36.13 PARK10 ? ? (Iceland) AR 1p32 PARK11 GIGYF2 GRB10-interacting GYF protein 2 AD 2q37.1 PARK12 ? ? X-R Xq21-q25 PARK13 HTRA2 serine protease AD 2p13.1 PARK14 PLA2G6 phospholipase A2 (INAD) AR 22q13.1 PARK15 FBXO7 F-box only protein 7 AR 22q12.3 PARK16 ? Discovered by GWAS ? 1q32 PARK17 VPS35 vacuolar protein sorting 35 AD 16q11.2 PARK18 EIF4G1 initiation of protein synth AD 3q27.1 PARK19 DNAJC6 auxilin AR 1p31.3 PARK20 SYNJ1 synaptojanin 1 AR 21q22.11 PARK21 DNAJC13 8/RME-8 AD 3q22.1 PARK22 CHCHD2 AD 7p11.2 PARK23 VPS13C AR 15q22 Gene Localizations Identified in PD Disorder Symbol Protein Transmission Chromosome PD GBA β-glucocerebrosidase AD 1q21 SCA2 -

Targeting Fibrosis in the Duchenne Muscular Dystrophy Mice Model: an Uphill Battle
bioRxiv preprint doi: https://doi.org/10.1101/2021.01.20.427485; this version posted January 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Title: Targeting fibrosis in the Duchenne Muscular Dystrophy mice model: an uphill battle 2 Marine Theret1#, Marcela Low1#, Lucas Rempel1, Fang Fang Li1, Lin Wei Tung1, Osvaldo 3 Contreras3,4, Chih-Kai Chang1, Andrew Wu1, Hesham Soliman1,2, Fabio M.V. Rossi1 4 1School of Biomedical Engineering and the Biomedical Research Centre, Department of Medical 5 Genetics, 2222 Health Sciences Mall, Vancouver, BC, V6T 1Z3, Canada 6 2Department of Pharmacology and Toxicology, Faculty of Pharmaceutical Sciences, Minia 7 University, Minia, Egypt 8 3Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, 9 Darlinghurst, NSW, 2010, Australia 10 4Departamento de Biología Celular y Molecular and Center for Aging and Regeneration (CARE- 11 ChileUC), Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, 8331150 12 Santiago, Chile 13 # Denotes Co-first authorship 14 15 Keywords: drug screening, fibro/adipogenic progenitors, fibrosis, repair, skeletal muscle. 16 Correspondence to: 17 Marine Theret 18 School of Biomedical Engineering and the Biomedical Research Centre 19 University of British Columbia 20 2222 Health Sciences Mall, Vancouver, British Columbia 21 Tel: +1(604) 822 0441 fax: +1(604) 822 7815 22 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.01.20.427485; this version posted January 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Identification and Characterization of TPRKB Dependency in TP53 Deficient Cancers
Identification and Characterization of TPRKB Dependency in TP53 Deficient Cancers. by Kelly Kennaley A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Molecular and Cellular Pathology) in the University of Michigan 2019 Doctoral Committee: Associate Professor Zaneta Nikolovska-Coleska, Co-Chair Adjunct Associate Professor Scott A. Tomlins, Co-Chair Associate Professor Eric R. Fearon Associate Professor Alexey I. Nesvizhskii Kelly R. Kennaley [email protected] ORCID iD: 0000-0003-2439-9020 © Kelly R. Kennaley 2019 Acknowledgements I have immeasurable gratitude for the unwavering support and guidance I received throughout my dissertation. First and foremost, I would like to thank my thesis advisor and mentor Dr. Scott Tomlins for entrusting me with a challenging, interesting, and impactful project. He taught me how to drive a project forward through set-backs, ask the important questions, and always consider the impact of my work. I’m truly appreciative for his commitment to ensuring that I would get the most from my graduate education. I am also grateful to the many members of the Tomlins lab that made it the supportive, collaborative, and educational environment that it was. I would like to give special thanks to those I’ve worked closely with on this project, particularly Dr. Moloy Goswami for his mentorship, Lei Lucy Wang, Dr. Sumin Han, and undergraduate students Bhavneet Singh, Travis Weiss, and Myles Barlow. I am also grateful for the support of my thesis committee, Dr. Eric Fearon, Dr. Alexey Nesvizhskii, and my co-mentor Dr. Zaneta Nikolovska-Coleska, who have offered guidance and critical evaluation since project inception. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Integrated Transcriptome and Methylome Analysis in Youth at High Risk for Bipolar Disorder: a Preliminary Analysis
OPEN Citation: Transl Psychiatry (2017) 7, e1059; doi:10.1038/tp.2017.32 www.nature.com/tp ORIGINAL ARTICLE Integrated transcriptome and methylome analysis in youth at high risk for bipolar disorder: a preliminary analysis GR Fries1, J Quevedo1,2,3,4, CP Zeni2, IF Kazimi2, G Zunta-Soares2, DE Spiker2, CL Bowden5, C Walss-Bass1,2,3 and JC Soares1,2 First-degree relatives of patients with bipolar disorder (BD), particularly their offspring, have a higher risk of developing BD and other mental illnesses than the general population. However, the biological mechanisms underlying this increased risk are still unknown, particularly because most of the studies so far have been conducted in chronically ill adults and not in unaffected youth at high risk. In this preliminary study we analyzed genome-wide expression and methylation levels in peripheral blood mononuclear cells from children and adolescents from three matched groups: BD patients, unaffected offspring of bipolar parents (high risk) and controls (low risk). By integrating gene expression and DNA methylation and comparing the lists of differentially expressed genes and differentially methylated probes between groups, we were able to identify 43 risk genes that discriminate patients and high-risk youth from controls. Pathway analysis showed an enrichment of the glucocorticoid receptor (GR) pathway with the genes MED1, HSPA1L, GTF2A1 and TAF15, which might underlie the previously reported role of stress response in the risk for BD in vulnerable populations. Cell-based assays indicate a GR hyporesponsiveness in cells from adult BD patients compared to controls and suggest that these GR-related genes can be modulated by DNA methylation, which poses the theoretical possibility of manipulating their expression as a means to counteract the familial risk presented by those subjects. -

Human Kinome Profiling Identifies a Requirement for AMP-Activated
Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection Laura J. Terrya, Livia Vastagb,1, Joshua D. Rabinowitzb, and Thomas Shenka,2 aDepartment of Molecular Biology and bDepartment of Chemistry and the Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ 08544 Contributed by Thomas Shenk, January 11, 2012 (sent for review December 29, 2011) Human cytomegalovirus (HCMV) modulates numerous cellular (7). Thus, the connections between AMPK activity and metabolic signaling pathways. Alterations in signaling are evident from the changes during HCMV infection have remained unclear. broad changes in cellular phosphorylation that occur during HCMV We confirmed the requirement for AMPK during infection, infection and from the altered activity of multiple kinases. Here we and we show that an AMPK antagonist, compound C, blocks report a comprehensive RNAi screen, which predicts that 106 cellular HCMV-induced changes to glycolysis and inhibits viral gene kinases influence growth of the virus, most of which were not expression. These studies argue that AMPK or a related, com- previously linked to HCMV replication. Multiple elements of the pound C-sensitive kinase is an essential contributor to metabolic AMP-activated protein kinase (AMPK) pathway scored in the screen. changes initiated by HCMV and provide unique insight into As a regulator of carbon and nucleotide metabolism, AMPK is poised potential antiviral strategies. to activate many of the metabolic pathways induced by HCMV infection. An AMPK inhibitor, compound C, blocked a substantial Results portion of HCMV-induced metabolic changes, inhibited the accumu- HumanKinomeScreenIdentifies Putative Effectors of HCMV Replication. lation of all HCMV proteins tested, and markedly reduced the We conducted an siRNA screen of the human kinome to perform an production of infectious progeny. -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

The Hsp70/Hsp90 Chaperone Machinery in Neurodegenerative Diseases
REVIEW published: 16 May 2017 doi: 10.3389/fnins.2017.00254 The Hsp70/Hsp90 Chaperone Machinery in Neurodegenerative Diseases Rachel E. Lackie 1, 2, Andrzej Maciejewski 1, 3, Valeriy G. Ostapchenko 1, Jose Marques-Lopes 1, Wing-Yiu Choy 3, Martin L. Duennwald 4, Vania F. Prado 1, 2, 5, 6 and Marco A. M. Prado 1, 2, 5, 6* 1 Molecular Medicine, Robarts Research Institute, University of Western Ontario, London, ON, Canada, 2 Program in Neuroscience, University of Western Ontario, London, ON, Canada, 3 Department of Biochemistry, University of Western Ontario, London, ON, Canada, 4 Department of Pathology and Laboratory Medicine, University of Western Ontario, London, ON, Canada, 5 Department of Physiology and Pharmacology, University of Western Ontario, London, ON, Canada, 6 Department of Anatomy and Cell Biology, Schulich School of Medicine and Dentistry, University of Western Ontario, London, ON, Canada The accumulation of misfolded proteins in the human brain is one of the critical features of many neurodegenerative diseases, including Alzheimer’s disease (AD). Assembles of beta-amyloid (Aβ) peptide—either soluble (oligomers) or insoluble (plaques) and Edited by: Cintia Roodveldt, of tau protein, which form neurofibrillary tangles, are the major hallmarks of AD. Centro Andaluz de Biología Molecular Chaperones and co-chaperones regulate protein folding and client maturation, but they y Medicina Regenerativa, Spain also target misfolded or aggregated proteins for refolding or for degradation, mostly Reviewed by: Toshihide Takeuchi, by the proteasome. They form an important line of defense against misfolded proteins Osaka University, Japan and are part of the cellular quality control system. The heat shock protein (Hsp) family, Janine Kirstein, particularly Hsp70 and Hsp90, plays a major part in this process and it is well-known to Leibniz Institute for Molecular Pharmacology (FMP), Germany regulate protein misfolding in a variety of diseases, including tau levels and toxicity in AD. -
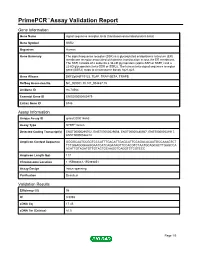
Download Validation Data
PrimePCR™Assay Validation Report Gene Information Gene Name signal sequence receptor, beta (translocon-associated protein beta) Gene Symbol SSR2 Organism Human Gene Summary The signal sequence receptor (SSR) is a glycosylated endoplasmic reticulum (ER) membrane receptor associated with protein translocation across the ER membrane. The SSR consists of 2 subunits a 34-kD glycoprotein (alpha-SSR or SSR1) and a 22-kD glycoprotein (beta-SSR or SSR2). The human beta-signal sequence receptor gene (SSR2) maps to chromosome bands 1q21-q23. Gene Aliases DKFZp686F19123, TLAP, TRAP-BETA, TRAPB RefSeq Accession No. NC_000001.10, NT_004487.19 UniGene ID Hs.74564 Ensembl Gene ID ENSG00000163479 Entrez Gene ID 6746 Assay Information Unique Assay ID qHsaCID0014663 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000295702, ENST00000529008, ENST00000480567, ENST00000531917, ENST00000526212 Amplicon Context Sequence GGGGCAATCCGGTCCCATTTGACATTGAGCATTCCAGACACAATGCCAAAGTCT TCTGGAGGGAAGGAATCATCAGATAGTTCCACGTCTAATGCAGCACTTGAGCCA ACATTGTAGATGTTGTACTGCAAGGTCAGGTCTCGTCCC Amplicon Length (bp) 117 Chromosome Location 1:155988061-155989851 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9998 cDNA Cq 17.45 cDNA Tm (Celsius) 81.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report SSR2, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference -

CLN8 Mutations Presenting with a Phenotypic Continuum of Neuronal Ceroid Lipofuscinosis—Literature Review and Case Report
G C A T T A C G G C A T genes Article CLN8 Mutations Presenting with a Phenotypic Continuum of Neuronal Ceroid Lipofuscinosis—Literature Review and Case Report Magdalena Badura-Stronka 1,*,†, Anna Winczewska-Wiktor 2,†, Anna Pietrzak 3,†, Adam Sebastian Hirschfeld 1, Tomasz Zemojtel 4, Katarzyna Woły ´nska 1, Katarzyna Bednarek-Rajewska 5, Monika Seget-Dubaniewicz 5, Agnieszka Matheisel 6, Anna Latos-Bielenska 1 and Barbara Steinborn 2 1 Chair and Department of Medical Genetics, Poznan University of Medical Sciences, 60-352 Poznan, Poland; [email protected] (A.S.H.); [email protected] (K.W.); [email protected] (A.L.-B.) 2 Chair and Department of Developmental Neurology, Poznan University of Medical Sciences, 60-355 Poznan, Poland; [email protected] (A.W.-W.); [email protected] (B.S.) 3 Department of Neurology, 10th Military Research Hospital and Polyclinic, 85-681 Bydgoszcz, Poland; [email protected] 4 BIH Genomics Core Unit, Campus Mitte, Charite University Medicine, 13353 Berlin, Germany; [email protected] 5 Department of Clinical Pathology, Poznan University of Medical Sciences, 60-355 Poznan, Poland; [email protected] (K.B.-R.); [email protected] (M.S.-D.) 6 Citation: Badura-Stronka, M.; Department of Developmental Neurology, Gdansk Medical University, 80-307 Gdansk, Poland; Winczewska-Wiktor, A.; Pietrzak, A.; [email protected] * Correspondence: [email protected] Hirschfeld, A.S.; Zemojtel, T.; † These authors contributed equally to this work. Woły´nska,K.; Bednarek-Rajewska, K.; Seget-Dubaniewicz, M.; Matheisel, A.; Latos-Bielenska, A.; Steinborn, B. Abstract: CLN8 is a ubiquitously expressed membrane-spanning protein that localizes primarily CLN8 Mutations Presenting with a in the ER, with partial localization in the ER-Golgi intermediate compartment.