PDF-Document
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Variation Across the Human Olfactory Receptor Repertoire Alters Odor Perception
bioRxiv preprint doi: https://doi.org/10.1101/212431; this version posted November 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genetic variation across the human olfactory receptor repertoire alters odor perception Casey Trimmer1,*, Andreas Keller2, Nicolle R. Murphy1, Lindsey L. Snyder1, Jason R. Willer3, Maira Nagai4,5, Nicholas Katsanis3, Leslie B. Vosshall2,6,7, Hiroaki Matsunami4,8, and Joel D. Mainland1,9 1Monell Chemical Senses Center, Philadelphia, Pennsylvania, USA 2Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, New York, USA 3Center for Human Disease Modeling, Duke University Medical Center, Durham, North Carolina, USA 4Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina, USA 5Department of Biochemistry, University of Sao Paulo, Sao Paulo, Brazil 6Howard Hughes Medical Institute, New York, New York, USA 7Kavli Neural Systems Institute, New York, New York, USA 8Department of Neurobiology and Duke Institute for Brain Sciences, Duke University Medical Center, Durham, North Carolina, USA 9Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA *[email protected] ABSTRACT The human olfactory receptor repertoire is characterized by an abundance of genetic variation that affects receptor response, but the perceptual effects of this variation are unclear. To address this issue, we sequenced the OR repertoire in 332 individuals and examined the relationship between genetic variation and 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Structural Capacitance in Protein Evolution and Human Diseases
Article Structural Capacitance in Protein Evolution and Human Diseases Chen Li 1,2, Liah V.T. Clark 1, Rory Zhang 1, Benjamin T. Porebski 1,3, Julia M. McCoey 1, Natalie A. Borg 1, Geoffrey I. Webb 4, Itamar Kass 1,5, Malcolm Buckle 6, Jiangning Song 1, Adrian Woolfson 7 and Ashley M. Buckle 1 1 - Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria 3800, Australia 2 - Department of Biology, Institute of Molecular Systems Biology, ETH Zurich, Zurich 8093, Switzerland 3 - Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge, CB2 0QH, UK 4 - Faculty of Information Technology, Monash University, Clayton, Victoria 3800, Australia 5 - Amai Proteins, Prof. A. D. Bergman 2B, Suite 212, Rehovot 7670504, Israel 6 - LBPA, ENS Cachan, CNRS, Université Paris-Saclay, F-94235 Cachan, France 7 - Nouscom, Baumleingasse, CH-4051 Basel, Switzerland Correspondence to Adrian Woolfson and Ashley M. Buckle: [email protected]; [email protected] https://doi.org/10.1016/j.jmb.2018.06.051 Edited by Dan Tawfik Abstract Canonical mechanisms of protein evolution include the duplication and diversification of pre-existing folds through genetic alterations that include point mutations, insertions, deletions, and copy number amplifications, as well as post-translational modifications that modify processes such as folding efficiency and cellular localization. Following a survey of the human mutation database, we have identified an additional mechanism that we term “structural capacitance,” which results in the de novo generation of microstructure in previously disordered regions. We suggest that the potential for structural capacitance confers select proteins with the capacity to evolve over rapid timescales, facilitating saltatory evolution as opposed to gradualistic canonical Darwinian mechanisms. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Structural Capacitance in Protein Evolution and Human Diseases
Structural Capacitance in Protein Evolution and Human Diseases Adrian Woolfson, Ashley Buckle, Natalie Borg, Geoffrey Webb, Itamar Kass, Malcolm Buckle, Jiangning Song, Chen Li, Liah Clark, Rory Zhang, et al. To cite this version: Adrian Woolfson, Ashley Buckle, Natalie Borg, Geoffrey Webb, Itamar Kass, et al.. Structural Ca- pacitance in Protein Evolution and Human Diseases. Journal of Molecular Biology, Elsevier, 2018, 10.1016/j.jmb.2018.06.051. hal-02368321 HAL Id: hal-02368321 https://hal.archives-ouvertes.fr/hal-02368321 Submitted on 20 Nov 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Structural Capacitance in Protein Evolution and Human Diseases Chen Li, Liah Clark, Rory Zhang, Benjamin Porebski, Julia Mccoey, Natalie Borg, Geoffrey Webb, Itamar Kass, Malcolm Buckle, Jiangning Song, etal. To cite this version: Chen Li, Liah Clark, Rory Zhang, Benjamin Porebski, Julia Mccoey, et al.. Structural Capaci- tance in Protein Evolution and Human Diseases. Journal of Molecular Biology, Elsevier, 2018, 10.1016/j.jmb.2018.06.051. hal-02368321 HAL Id: hal-02368321 https://hal.archives-ouvertes.fr/hal-02368321 Submitted on 20 Nov 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. -

Antibody List
產品編號 產品名稱 PA569955 1110059E24Rik Polyclonal Antibody PA569956 1110059E24Rik Polyclonal Antibody PA570131 1190002N15Rik Polyclonal Antibody 01-1234-42 123count eBeads Counting Beads MA512242 14.3.3 Pan Monoclonal Antibody (CG15) LFMA0074 14-3-3 beta Monoclonal Antibody (60C10) LFPA0077 14-3-3 beta Polyclonal Antibody PA137002 14-3-3 beta Polyclonal Antibody PA14647 14-3-3 beta Polyclonal Antibody PA515477 14-3-3 beta Polyclonal Antibody PA517425 14-3-3 beta Polyclonal Antibody PA522264 14-3-3 beta Polyclonal Antibody PA529689 14-3-3 beta Polyclonal Antibody MA134561 14-3-3 beta/epsilon/zeta Monoclonal Antibody (3C8) MA125492 14-3-3 beta/zeta Monoclonal Antibody (22-IID8B) MA125665 14-3-3 beta/zeta Monoclonal Antibody (4E2) 702477 14-3-3 delta/zeta Antibody (1H9L19), ABfinity Rabbit Monoclonal 711507 14-3-3 delta/zeta Antibody (1HCLC), ABfinity Rabbit Oligoclonal 702241 14-3-3 epsilon Antibody (5H10L5), ABfinity Rabbit Monoclonal 711273 14-3-3 epsilon Antibody (5HCLC), ABfinity Rabbit Oligoclonal PA517104 14-3-3 epsilon Polyclonal Antibody PA528937 14-3-3 epsilon Polyclonal Antibody PA529773 14-3-3 epsilon Polyclonal Antibody PA575298 14-3-3 eta (Lys81) Polyclonal Antibody MA524792 14-3-3 eta Monoclonal Antibody PA528113 14-3-3 eta Polyclonal Antibody PA529774 14-3-3 eta Polyclonal Antibody PA546811 14-3-3 eta Polyclonal Antibody MA116588 14-3-3 gamma Monoclonal Antibody (HS23) MA116587 14-3-3 gamma Monoclonal Antibody (KC21) PA529690 14-3-3 gamma Polyclonal Antibody PA578233 14-3-3 gamma Polyclonal Antibody 510700 14-3-3 Pan Polyclonal -

Misexpression of Cancer/Testis (Ct) Genes in Tumor Cells and the Potential Role of Dream Complex and the Retinoblastoma Protein Rb in Soma-To-Germline Transformation
Michigan Technological University Digital Commons @ Michigan Tech Dissertations, Master's Theses and Master's Reports 2019 MISEXPRESSION OF CANCER/TESTIS (CT) GENES IN TUMOR CELLS AND THE POTENTIAL ROLE OF DREAM COMPLEX AND THE RETINOBLASTOMA PROTEIN RB IN SOMA-TO-GERMLINE TRANSFORMATION SABHA M. ALHEWAT Michigan Technological University, [email protected] Copyright 2019 SABHA M. ALHEWAT Recommended Citation ALHEWAT, SABHA M., "MISEXPRESSION OF CANCER/TESTIS (CT) GENES IN TUMOR CELLS AND THE POTENTIAL ROLE OF DREAM COMPLEX AND THE RETINOBLASTOMA PROTEIN RB IN SOMA-TO- GERMLINE TRANSFORMATION", Open Access Master's Thesis, Michigan Technological University, 2019. https://doi.org/10.37099/mtu.dc.etdr/933 Follow this and additional works at: https://digitalcommons.mtu.edu/etdr Part of the Cancer Biology Commons, and the Cell Biology Commons MISEXPRESSION OF CANCER/TESTIS (CT) GENES IN TUMOR CELLS AND THE POTENTIAL ROLE OF DREAM COMPLEX AND THE RETINOBLASTOMA PROTEIN RB IN SOMA-TO-GERMLINE TRANSFORMATION By Sabha Salem Alhewati A THESIS Submitted in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE In Biological Sciences MICHIGAN TECHNOLOGICAL UNIVERSITY 2019 © 2019 Sabha Alhewati This thesis has been approved in partial fulfillment of the requirements for the Degree of MASTER OF SCIENCE in Biological Sciences. Department of Biological Sciences Thesis Advisor: Paul Goetsch. Committee Member: Ebenezer Tumban. Committee Member: Zhiying Shan. Department Chair: Chandrashekhar Joshi. Table of Contents List of figures .......................................................................................................................v -

Supplementary Information
Supplementary Information Structural Capacitance in Protein Evolution and Human Diseases Chen Li, Liah V T Clark, Rory Zhang, Benjamin T Porebski, Julia M. McCoey, Natalie A. Borg, Geoffrey I. Webb, Itamar Kass, Malcolm Buckle, Jiangning Song, Adrian Woolfson, and Ashley M. Buckle Supplementary tables Table S1. Disorder prediction using the human disease and polymorphisms dataseta OR DR OO OD DD DO mutations mutations 24,758 650 2,741 513 Disease 25,408 3,254 97.44% 2.56% 84.23% 15.77% 26,559 809 11,135 1,218 Non-disease 27,368 12,353 97.04% 2.96% 90.14% 9.86% ahttp://www.uniprot.org/docs/humsavar [1] (see Materials and Methdos). The numbers listed are the ones of unique mutations. ‘Unclassifiied’ mutations, according to the UniProt, were not counted. O = predicted as ordered; OR = Ordered regions D = predicted as disordered; DR = Disordered regions 1 Table S2. Mutations in long disordered regions (LDRs) of human proteins predicted to produce a DO transitiona Average # disorder # disorder # disorder # order UniProt/dbSNP Protein Mutation Disease length of predictors predictors predictorsb predictorsc LDRd in D2P2e for LDRf UHRF1-binding protein 1- A0JNW5/rs7296162 like S1147L - 4 2^ 101 6 3 A4D1E1/rs801841 Zinc finger protein 804B V1195I - 3* 2^ 37 6 1 A6NJV1/rs2272466 UPF0573 protein C2orf70 Q177L - 2* 4 34 3 1 Golgin subfamily A member A7E2F4/rs347880 8A K480N - 2* 2^ 91 N/A 2 Axonemal dynein light O14645/rs11749 intermediate polypeptide 1 A65V - 3* 3 43 N/A 2 Centrosomal protein of 290 O15078/rs374852145 kDa R2210C - 2 3 123 5 1 Fanconi -

Single Cell Derived Clonal Analysis of Human Glioblastoma Links
SUPPLEMENTARY INFORMATION: Single cell derived clonal analysis of human glioblastoma links functional and genomic heterogeneity ! Mona Meyer*, Jüri Reimand*, Xiaoyang Lan, Renee Head, Xueming Zhu, Michelle Kushida, Jane Bayani, Jessica C. Pressey, Anath Lionel, Ian D. Clarke, Michael Cusimano, Jeremy Squire, Stephen Scherer, Mark Bernstein, Melanie A. Woodin, Gary D. Bader**, and Peter B. Dirks**! ! * These authors contributed equally to this work.! ** Correspondence: [email protected] or [email protected]! ! Supplementary information - Meyer, Reimand et al. Supplementary methods" 4" Patient samples and fluorescence activated cell sorting (FACS)! 4! Differentiation! 4! Immunocytochemistry and EdU Imaging! 4! Proliferation! 5! Western blotting ! 5! Temozolomide treatment! 5! NCI drug library screen! 6! Orthotopic injections! 6! Immunohistochemistry on tumor sections! 6! Promoter methylation of MGMT! 6! Fluorescence in situ Hybridization (FISH)! 7! SNP6 microarray analysis and genome segmentation! 7! Calling copy number alterations! 8! Mapping altered genome segments to genes! 8! Recurrently altered genes with clonal variability! 9! Global analyses of copy number alterations! 9! Phylogenetic analysis of copy number alterations! 10! Microarray analysis! 10! Gene expression differences of TMZ resistant and sensitive clones of GBM-482! 10! Reverse transcription-PCR analyses! 11! Tumor subtype analysis of TMZ-sensitive and resistant clones! 11! Pathway analysis of gene expression in the TMZ-sensitive clone of GBM-482! 11! Supplementary figures and tables" 13" "2 Supplementary information - Meyer, Reimand et al. Table S1: Individual clones from all patient tumors are tumorigenic. ! 14! Fig. S1: clonal tumorigenicity.! 15! Fig. S2: clonal heterogeneity of EGFR and PTEN expression.! 20! Fig. S3: clonal heterogeneity of proliferation.! 21! Fig. -

Integrated Epigenomic Analysis Stratifies Chromatin Remodellers Into
Giles et al. Epigenetics & Chromatin (2019) 12:12 https://doi.org/10.1186/s13072-019-0258-9 Epigenetics & Chromatin RESEARCH Open Access Integrated epigenomic analysis stratifes chromatin remodellers into distinct functional groups Katherine A. Giles1, Cathryn M. Gould1, Qian Du1, Ksenia Skvortsova1, Jenny Z. Song1, Madhavi P. Maddugoda1, Joanna Achinger‑Kawecka1,2, Clare Stirzaker1,2, Susan J. Clark1,2† and Phillippa C. Taberlay2,3*† Abstract Background: ATP‑dependent chromatin remodelling complexes are responsible for establishing and maintaining the positions of nucleosomes. Chromatin remodellers are targeted to chromatin by transcription factors and non‑ coding RNA to remodel the chromatin into functional states. However, the infuence of chromatin remodelling on shaping the functional epigenome is not well understood. Moreover, chromatin remodellers have not been exten‑ sively explored as a collective group across two‑dimensional and three‑dimensional epigenomic layers. Results: Here, we have integrated the genome‑wide binding profles of eight chromatin remodellers together with DNA methylation, nucleosome positioning, histone modifcation and Hi‑C chromosomal contacts to reveal that chro‑ matin remodellers can be stratifed into two functional groups. Group 1 (BRG1, SNF2H, CHD3 and CHD4) has a clear preference for binding at ‘actively marked’ chromatin and Group 2 (BRM, INO80, SNF2L and CHD1) for ‘repressively marked’ chromatin. We fnd that histone modifcations and chromatin architectural features, but not DNA methyla‑ tion, stratify the remodellers into these functional groups. Conclusions: Our fndings suggest that chromatin remodelling events are synchronous and that chromatin remod‑ ellers themselves should be considered simultaneously and not as individual entities in isolation or necessarily by structural similarity, as they are traditionally classifed. -
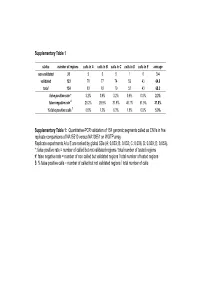
Quantitative-PCR Validation of 154 Genomic Segments Called As Cnvs in Five Replicat
Supplementary Table 1 status number of regions calls in A calls in B calls in C calls in D calls in E average non validated 31 5 6 5 1 0 3.4 validated 123 78 77 74 52 43 64.8 total 154 83 83 79 53 43 68.2 false positive rate * 3.2% 3.9% 3.2% 0.6% 0.0% 2.2% false negative rate # 29.2% 29.9% 31.8% 46.1% 51.9% 37.8% % false positive calls $ 6.0% 7.2% 6.3% 1.9% 0.0% 5.0% Supplementary Table 1: Quantitative-PCR validation of 154 genomic segments called as CNVs in five replicate comparisons of NA15510 versus NA10851 on WGTP array Replicate experiments A to E are ranked by global SDe (A: 0.033; B: 0.033; C: 0.036; D: 0.039; E: 0.053). *: false positive rate = number of called but not validated regions / total number of tested regions #: false negative rate = number of non called but validated regions / total number of tested regions $: % false positive calls = number of called but not validated regions / total number of calls False positive estimates for 500K EA CNV calls Total Rep1 Rep2 Rep3 Avg (unique) Validated 33 28 32 31 38 Not validated 2 2 2 2 5 Total 35 30 34 33 43 % False positive 5.71% 6.67% 5.88% 6.09% - % False negative 13.16% 26.32% 15.79% 18.42% - Supplementary Table 2A : Quantitative PCR validation of 43 unique CNV regions called as CNVs in three replicate comparisons of NA15510 versus NA10851 using the 500K EA array. -

Detection of H3k4me3 Identifies Neurohiv Signatures, Genomic
viruses Article Detection of H3K4me3 Identifies NeuroHIV Signatures, Genomic Effects of Methamphetamine and Addiction Pathways in Postmortem HIV+ Brain Specimens that Are Not Amenable to Transcriptome Analysis Liana Basova 1, Alexander Lindsey 1, Anne Marie McGovern 1, Ronald J. Ellis 2 and Maria Cecilia Garibaldi Marcondes 1,* 1 San Diego Biomedical Research Institute, San Diego, CA 92121, USA; [email protected] (L.B.); [email protected] (A.L.); [email protected] (A.M.M.) 2 Departments of Neurosciences and Psychiatry, University of California San Diego, San Diego, CA 92103, USA; [email protected] * Correspondence: [email protected] Abstract: Human postmortem specimens are extremely valuable resources for investigating trans- lational hypotheses. Tissue repositories collect clinically assessed specimens from people with and without HIV, including age, viral load, treatments, substance use patterns and cognitive functions. One challenge is the limited number of specimens suitable for transcriptional studies, mainly due to poor RNA quality resulting from long postmortem intervals. We hypothesized that epigenomic Citation: Basova, L.; Lindsey, A.; signatures would be more stable than RNA for assessing global changes associated with outcomes McGovern, A.M.; Ellis, R.J.; of interest. We found that H3K27Ac or RNA Polymerase (Pol) were not consistently detected by Marcondes, M.C.G. Detection of H3K4me3 Identifies NeuroHIV Chromatin Immunoprecipitation (ChIP), while the enhancer H3K4me3 histone modification was Signatures, Genomic Effects of abundant and stable up to the 72 h postmortem. We tested our ability to use H3K4me3 in human Methamphetamine and Addiction prefrontal cortex from HIV+ individuals meeting criteria for methamphetamine use disorder or not Pathways in Postmortem HIV+ Brain (Meth +/−) which exhibited poor RNA quality and were not suitable for transcriptional profiling.