Supplemental Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Variation Across the Human Olfactory Receptor Repertoire Alters Odor Perception
bioRxiv preprint doi: https://doi.org/10.1101/212431; this version posted November 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genetic variation across the human olfactory receptor repertoire alters odor perception Casey Trimmer1,*, Andreas Keller2, Nicolle R. Murphy1, Lindsey L. Snyder1, Jason R. Willer3, Maira Nagai4,5, Nicholas Katsanis3, Leslie B. Vosshall2,6,7, Hiroaki Matsunami4,8, and Joel D. Mainland1,9 1Monell Chemical Senses Center, Philadelphia, Pennsylvania, USA 2Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, New York, USA 3Center for Human Disease Modeling, Duke University Medical Center, Durham, North Carolina, USA 4Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina, USA 5Department of Biochemistry, University of Sao Paulo, Sao Paulo, Brazil 6Howard Hughes Medical Institute, New York, New York, USA 7Kavli Neural Systems Institute, New York, New York, USA 8Department of Neurobiology and Duke Institute for Brain Sciences, Duke University Medical Center, Durham, North Carolina, USA 9Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA *[email protected] ABSTRACT The human olfactory receptor repertoire is characterized by an abundance of genetic variation that affects receptor response, but the perceptual effects of this variation are unclear. To address this issue, we sequenced the OR repertoire in 332 individuals and examined the relationship between genetic variation and 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. -

Whole Exome Sequencing in Families at High Risk for Hodgkin Lymphoma: Identification of a Predisposing Mutation in the KDR Gene
Hodgkin Lymphoma SUPPLEMENTARY APPENDIX Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene Melissa Rotunno, 1 Mary L. McMaster, 1 Joseph Boland, 2 Sara Bass, 2 Xijun Zhang, 2 Laurie Burdett, 2 Belynda Hicks, 2 Sarangan Ravichandran, 3 Brian T. Luke, 3 Meredith Yeager, 2 Laura Fontaine, 4 Paula L. Hyland, 1 Alisa M. Goldstein, 1 NCI DCEG Cancer Sequencing Working Group, NCI DCEG Cancer Genomics Research Laboratory, Stephen J. Chanock, 5 Neil E. Caporaso, 1 Margaret A. Tucker, 6 and Lynn R. Goldin 1 1Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 2Cancer Genomics Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 3Ad - vanced Biomedical Computing Center, Leidos Biomedical Research Inc.; Frederick National Laboratory for Cancer Research, Frederick, MD; 4Westat, Inc., Rockville MD; 5Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; and 6Human Genetics Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA ©2016 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2015.135475 Received: August 19, 2015. Accepted: January 7, 2016. Pre-published: June 13, 2016. Correspondence: [email protected] Supplemental Author Information: NCI DCEG Cancer Sequencing Working Group: Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Anand Pathak, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, Guoqin Yu NCI DCEG Cancer Genomics Research Laboratory: Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. -

WO 2019/068007 Al Figure 2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/068007 Al 04 April 2019 (04.04.2019) W 1P O PCT (51) International Patent Classification: (72) Inventors; and C12N 15/10 (2006.01) C07K 16/28 (2006.01) (71) Applicants: GROSS, Gideon [EVIL]; IE-1-5 Address C12N 5/10 (2006.0 1) C12Q 1/6809 (20 18.0 1) M.P. Korazim, 1292200 Moshav Almagor (IL). GIBSON, C07K 14/705 (2006.01) A61P 35/00 (2006.01) Will [US/US]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., C07K 14/725 (2006.01) P.O. Box 4044, 7403635 Ness Ziona (TL). DAHARY, Dvir [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (21) International Application Number: Box 4044, 7403635 Ness Ziona (IL). BEIMAN, Merav PCT/US2018/053583 [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (22) International Filing Date: Box 4044, 7403635 Ness Ziona (E.). 28 September 2018 (28.09.2018) (74) Agent: MACDOUGALL, Christina, A. et al; Morgan, (25) Filing Language: English Lewis & Bockius LLP, One Market, Spear Tower, SanFran- cisco, CA 94105 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of national protection available): AE, AG, AL, AM, 62/564,454 28 September 2017 (28.09.2017) US AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/649,429 28 March 2018 (28.03.2018) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (71) Applicant: IMMP ACT-BIO LTD. -

Predicting Human Olfactory Perception from Activities of Odorant Receptors
iScience ll OPEN ACCESS Article Predicting Human Olfactory Perception from Activities of Odorant Receptors Joel Kowalewski, Anandasankar Ray [email protected] odor perception HIGHLIGHTS Machine learning predicted activity of 34 human ORs for ~0.5 million chemicals chemical structure Activities of human ORs predicts OR activity could predict odor character using machine learning Few OR activities were needed to optimize r predictions of each odor e t c percept a AI r a odorant activates mul- h Behavior predictions in c Drosophila also need few r tiple ORs o olfactory receptor d o activities ts ic ed pr ity tiv ac OR Kowalewski & Ray, iScience 23, 101361 August 21, 2020 ª 2020 The Author(s). https://doi.org/10.1016/ j.isci.2020.101361 iScience ll OPEN ACCESS Article Predicting Human Olfactory Perception from Activities of Odorant Receptors Joel Kowalewski1 and Anandasankar Ray1,2,3,* SUMMARY Odor perception in humans is initiated by activation of odorant receptors (ORs) in the nose. However, the ORs linked to specific olfactory percepts are unknown, unlike in vision or taste where receptors are linked to perception of different colors and tastes. The large family of ORs (~400) and multiple receptors activated by an odorant present serious challenges. Here, we first use machine learning to screen ~0.5 million compounds for new ligands and identify enriched structural motifs for ligands of 34 human ORs. We next demonstrate that the activity of ORs successfully predicts many of the 146 different perceptual qualities of chem- icals. Although chemical features have been used to model odor percepts, we show that biologically relevant OR activity is often superior. -

Characterizing Genomic Duplication in Autism Spectrum Disorder by Edward James Higginbotham a Thesis Submitted in Conformity
Characterizing Genomic Duplication in Autism Spectrum Disorder by Edward James Higginbotham A thesis submitted in conformity with the requirements for the degree of Master of Science Graduate Department of Molecular Genetics University of Toronto © Copyright by Edward James Higginbotham 2020 i Abstract Characterizing Genomic Duplication in Autism Spectrum Disorder Edward James Higginbotham Master of Science Graduate Department of Molecular Genetics University of Toronto 2020 Duplication, the gain of additional copies of genomic material relative to its ancestral diploid state is yet to achieve full appreciation for its role in human traits and disease. Challenges include accurately genotyping, annotating, and characterizing the properties of duplications, and resolving duplication mechanisms. Whole genome sequencing, in principle, should enable accurate detection of duplications in a single experiment. This thesis makes use of the technology to catalogue disease relevant duplications in the genomes of 2,739 individuals with Autism Spectrum Disorder (ASD) who enrolled in the Autism Speaks MSSNG Project. Fine-mapping the breakpoint junctions of 259 ASD-relevant duplications identified 34 (13.1%) variants with complex genomic structures as well as tandem (193/259, 74.5%) and NAHR- mediated (6/259, 2.3%) duplications. As whole genome sequencing-based studies expand in scale and reach, a continued focus on generating high-quality, standardized duplication data will be prerequisite to addressing their associated biological mechanisms. ii Acknowledgements I thank Dr. Stephen Scherer for his leadership par excellence, his generosity, and for giving me a chance. I am grateful for his investment and the opportunities afforded me, from which I have learned and benefited. I would next thank Drs. -
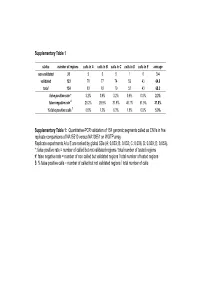
Quantitative-PCR Validation of 154 Genomic Segments Called As Cnvs in Five Replicat
Supplementary Table 1 status number of regions calls in A calls in B calls in C calls in D calls in E average non validated 31 5 6 5 1 0 3.4 validated 123 78 77 74 52 43 64.8 total 154 83 83 79 53 43 68.2 false positive rate * 3.2% 3.9% 3.2% 0.6% 0.0% 2.2% false negative rate # 29.2% 29.9% 31.8% 46.1% 51.9% 37.8% % false positive calls $ 6.0% 7.2% 6.3% 1.9% 0.0% 5.0% Supplementary Table 1: Quantitative-PCR validation of 154 genomic segments called as CNVs in five replicate comparisons of NA15510 versus NA10851 on WGTP array Replicate experiments A to E are ranked by global SDe (A: 0.033; B: 0.033; C: 0.036; D: 0.039; E: 0.053). *: false positive rate = number of called but not validated regions / total number of tested regions #: false negative rate = number of non called but validated regions / total number of tested regions $: % false positive calls = number of called but not validated regions / total number of calls False positive estimates for 500K EA CNV calls Total Rep1 Rep2 Rep3 Avg (unique) Validated 33 28 32 31 38 Not validated 2 2 2 2 5 Total 35 30 34 33 43 % False positive 5.71% 6.67% 5.88% 6.09% - % False negative 13.16% 26.32% 15.79% 18.42% - Supplementary Table 2A : Quantitative PCR validation of 43 unique CNV regions called as CNVs in three replicate comparisons of NA15510 versus NA10851 using the 500K EA array. -

Chr CNV Start CNV Stop Gene Gene Feature 1 37261312 37269719
chr CNV start CNV stop Gene Gene feature 1 37261312 37269719 Tmem131 closest upstream gene 1 37261312 37269719 Cnga3 closest downstream gene 1 41160869 41180390 Tmem182 closest upstream gene 1 41160869 41180390 2610017I09Rik closest downstream gene 1 66835123 66839616 1110028C15Rik in region 2 88714200 88719211 Olfr1206 closest upstream gene 2 88714200 88719211 Olfr1208 closest downstream gene 2 154840037 154846228 a in region 3 30065831 30417157 Mecom closest upstream gene 3 30065831 30417157 Arpm1 closest downstream gene 3 35476875 35495913 Sox2ot closest upstream gene 3 35476875 35495913 Atp11b closest downstream gene 3 39563408 39598697 Fat4 closest upstream gene 3 39563408 39598697 Intu closest downstream gene 3 94246481 94410611 Celf3 in region 3 94246481 94410611 Mrpl9 in region 3 94246481 94410611 Riiad1 in region 3 94246481 94410611 Snx27 in region 3 104311901 104319916 Lrig2 in region 3 144613709 144619149 Clca6 in region 3 144613709 144619149 Clca6 in region 4 108673 137301 Vmn1r2 closest downstream gene 4 3353037 5882883 6330407A03Rik in region 4 3353037 5882883 Chchd7 in region 4 3353037 5882883 Fam110b in region 4 3353037 5882883 Impad1 in region 4 3353037 5882883 Lyn in region 4 3353037 5882883 Mos in region 4 3353037 5882883 Penk in region 4 3353037 5882883 Plag1 in region 4 3353037 5882883 Rps20 in region 4 3353037 5882883 Sdr16c5 in region 4 3353037 5882883 Sdr16c6 in region 4 3353037 5882883 Tgs1 in region 4 3353037 5882883 Tmem68 in region 4 5919294 6304249 Cyp7a1 in region 4 5919294 6304249 Sdcbp in region 4 5919294 -

Clinical, Molecular, and Immune Analysis of Dabrafenib-Trametinib
Supplementary Online Content Chen G, McQuade JL, Panka DJ, et al. Clinical, molecular and immune analysis of dabrafenib-trametinib combination treatment for metastatic melanoma that progressed during BRAF inhibitor monotherapy: a phase 2 clinical trial. JAMA Oncology. Published online April 28, 2016. doi:10.1001/jamaoncol.2016.0509. eMethods. eReferences. eTable 1. Clinical efficacy eTable 2. Adverse events eTable 3. Correlation of baseline patient characteristics with treatment outcomes eTable 4. Patient responses and baseline IHC results eFigure 1. Kaplan-Meier analysis of overall survival eFigure 2. Correlation between IHC and RNAseq results eFigure 3. pPRAS40 expression and PFS eFigure 4. Baseline and treatment-induced changes in immune infiltrates eFigure 5. PD-L1 expression eTable 5. Nonsynonymous mutations detected by WES in baseline tumors This supplementary material has been provided by the authors to give readers additional information about their work. © 2016 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/30/2021 eMethods Whole exome sequencing Whole exome capture libraries for both tumor and normal samples were constructed using 100ng genomic DNA input and following the protocol as described by Fisher et al.,3 with the following adapter modification: Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter. In-solution hybrid selection was performed using the Illumina Rapid Capture Exome enrichment kit with 38Mb target territory (29Mb baited). The targeted region includes 98.3% of the intervals in the Refseq exome database. Dual-indexed libraries were pooled into groups of up to 96 samples prior to hybridization. -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Supplemental Table 1 (S1). the Chromosomal Regions and Genes Exhibiting Loss of Heterozygosity That Were Shared Between the HLRCC-Rccs of Both Patients 1 and 2
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Clin Pathol Supplemental Table 1 (S1). The chromosomal regions and genes exhibiting loss of heterozygosity that were shared between the HLRCC-RCCs of both Patients 1 and 2. Chromosome Position Genes 1 p13.1 ATP1A1, ATP1A1-AS1, LOC101929023, CD58, IGSF3, MIR320B1, C1orf137, CD2, PTGFRN, CD101, LOC101929099 1 p21.1* COL11A1, LOC101928436, RNPC3, AMY2B, ACTG1P4, AMY2A, AMY1A, AMY1C, AMY1B 1 p36.11 LOC101928728, ARID1A, PIGV, ZDHHC18, SFN, GPN2, GPATCH3, NR0B2, NUDC, KDF1, TRNP1, FAM46B, SLC9A1, WDTC1, TMEM222, ACTG1P20, SYTL1, MAP3K6, FCN3, CD164L2, GPR3, WASF2, AHDC1, FGR, IFI6 1 q21.3* KCNN3, PMVK, PBXIP1, PYGO2, LOC101928120, SHC1, CKS1B, MIR4258, FLAD1, LENEP, ZBTB7B, DCST2, DCST1, LOC100505666, ADAM15, EFNA4, EFNA3, EFNA1, SLC50A1, DPM3, KRTCAP2, TRIM46, MUC1, MIR92B, THBS3, MTX1, GBAP1, GBA, FAM189B, SCAMP3, CLK2, HCN3, PKLR, FDPS, RUSC1-AS1, RUSC1, ASH1L, MIR555, POU5F1P4, ASH1L-AS1, MSTO1, MSTO2P, YY1AP1, SCARNA26A, DAP3, GON4L, SCARNA26B, SYT11, RIT1, KIAA0907, SNORA80E, SCARNA4, RXFP4, ARHGEF2, MIR6738, SSR2, UBQLN4, LAMTOR2, RAB25, MEX3A, LMNA, SEMA4A, SLC25A44, PMF1, PMF1-BGLAP 1 q24.2–44* LOC101928650, GORAB, PRRX1, MROH9, FMO3, MIR1295A, MIR1295B, FMO6P, FMO2, FMO1, FMO4, TOP1P1, PRRC2C, MYOC, VAMP4, METTL13, DNM3, DNM3-IT1, DNM3OS, MIR214, MIR3120, MIR199A2, C1orf105, PIGC, SUCO, FASLG, TNFSF18, TNFSF4, LOC100506023, LOC101928673, -

Supplementary Data
A Suppl. Figure 1 A 1.0 p = 0.0192 0.8 0.6 0.4 Proportion Surviving 0.2 Gain Normal 0.0 0122436486072 Months After Esophagectomy B p = 0.034 Below Median Proportion Surviving Above Median Months After Esophagectomy Suppl. Figure 2 Suppl. Figure 3 Suppl. Figure 4 Suppl. Figure 5 A Score 0 Score 1 Score 2 Score 3 N=25, (22.5% of patients) N=53, (47.7% of patients) N=23, (20.7% of patients) N=10, (9.0% of patients) B 60 50 40 30 % of Patients 20 10 0 1 23 Score SUPPLEMENTARY MATERIAL Supplementary methods: Real-time PCR analysis. Real-time PCR analysis for CDK4 expression was performed in 108 tumors using Assays-on-Demand (AOD, Life technologies Corp, Carlsbad, CA). 88ng of cDNA (RNA equivalents) template was used in each qPCR reaction and assays were performed in triplicate. Quantification of CDK4 expression was measured relative to the geometric mean of five endogenous control genes (HMBS, POLRA, TBP, PGK, UCBH). Network-based analysis. Genes that were differently expressed between CDK6-positive and CDK6- negative samples with a p-value < 0.05 and fold-change >0.5 were identified using limma package from bioconductor (1). Network-based analysis was performed using the functional interaction (FI) network (2). Briefly, this consists of 10956 proteins and more than 200,000 curated functional interactions. We calculated pairwise shortest paths among genes of interest in the FI network, hierarchically clustered them based on the average linkage method, and then selected a cluster containing more than 80% altered genes. -

Apoptotic Cells Inflammasome Activity During the Uptake of Macrophage
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology published online 20 April 2012 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2012/04/20/jimmun ol.1103760 Complement Protein C1q Directs Macrophage Polarization and Limits Inflammasome Activity during the Uptake of Apoptotic Cells Marie E. Benoit, Elizabeth V. Clarke, Pedro Morgado, Deborah A. Fraser and Andrea J. Tenner J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/suppl/2012/04/20/jimmunol.110376 0.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. Published April 20, 2012, doi:10.4049/jimmunol.1103760 The Journal of Immunology Complement Protein C1q Directs Macrophage Polarization and Limits Inflammasome Activity during the Uptake of Apoptotic Cells Marie E. Benoit, Elizabeth V. Clarke, Pedro Morgado, Deborah A. Fraser, and Andrea J. Tenner Deficiency in C1q, the recognition component of the classical complement cascade and a pattern recognition receptor involved in apoptotic cell clearance, leads to lupus-like autoimmune diseases characterized by auto-antibodies to self proteins and aberrant innate immune cell activation likely due to impaired clearance of apoptotic cells.