On an Algorithmic Definition for the Components of the Minimal Cell
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
6 X 10.5 Long Title.P65
Cambridge University Press 978-0-521-51840-6 - An Introduction to Classical Nahuatl Michel Launey Excerpt More information Ī Ī Ī Ī Ī PART Ī Ī ONE Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī Ī © in this web service Cambridge University Press www.cambridge.org Cambridge University Press 978-0-521-51840-6 - An Introduction to Classical Nahuatl Michel Launey Excerpt More information PRELIMINARY LESSON Phonetics and Writing S ince the time of the conquest, Nahuatl has been written by means of theLatinalphabet.Thereis,therefore,alongtraditiontowhichitis preferable to conform for the most part. Nonetheless, for the following reasons, somewordsinthisbookarewritteninanorthographythatdiffersfromthe traditional one. ̈ Theorthographyis,ofcourse,“hispanicized.”Torepresentthephoneticele- ments of Nahuatl, the letters or combinations of letters that represent identical or similar sounds in Spanish are used. Hence, there is no problem with the sounds that exist in both languages, not to mention those that are lacking in Nahuatl (b, d, g, r etc.). On the other hand, those that exist in Nahuatl but not inSpanisharefoundinalternatespellings,orareevenaltogetherignored.In particular, this is the case with vowel length and (even worse) with the glottal stop (see Table 1.1), which are systematically marked only by two grammar- ians, Horacio Carochi and Aldama y Guevara, and in a text named Bancroft Dialogues.1 ̈ This defective character is heightened by a certain fluctuation because the orthography of Nahuatl has never really been fixed. Hence, certain texts represent the vowel /i/ indifferently with i or j, others always represent it with i but extend this spelling to consonantal /y/, that is, to a differ- ent phoneme. -

Saltillo Street Names Index: Autumn Gleen Dr
M R - 6 - E R R - 5 - E C c 1 2 3 D 4 5 6 7 U C S D A T R . E R D L 6 . 8 E 3 Y SNER ST. RD 2446 MES SALTILLO STREET NAMES INDEX: AUTUMN GLEEN DR. E3 OLD HWY. 51 E3 A BAUHAUS DR. E3 OLD NATCHEZ TRACE TRAIL G3 BENT GRASS CIR. H2 OLD SALTILLO RD. E4, F4, F5, G5, H4, H5 A BERCH TREE DR. F4 OLD SOUTH PLANTATION DR. E2 R D BIG HARPE TRAIL G2 OLIVIA CV. D4 BIRMINGHAM RIDGE RD. F1, F2, G3 7 PARKRIDGE DR, F4 8 3 BOLIVAR CIR. G3 PATTERSON CIR. E5 . D BOSTIC AVE. E4 PECAN DR. D5 R BRIDLE CV. E3 PETE AVE. F3 D U BRISCOE ST. E4 PEYTON LN. D4 H BROCK DR. F3 33 35 35 35 PINECREST ST. E4 ST 32 32 33 TOWERY . CARTWRIGHT AVE. E4 PINEHURST AVE. E4 31 36 31 R 4 3 D 3 2 CHAMPIONS CV. H2 PINEWOOD ST. E4 5 6 5 4 CHERRY HILL DR. C4 POPULARWOOD LN. F4 1 6 9 5 CHESTNUT LN. C4 PRINSTON DR. E3 1 CHICKASAW CIR. G3 PULLTIGHT RD. B4, C4 CHICKASAW TRAIL G3 PYLE DR. E4 CHRISTY WAY C4 QUAIL CREEK RD. E3 CO 653-B H2, H3 RAILROAD AVE. E5 CO 653-D G3 RD 189 F3 CO 2012 E2, E3 RD 251 D2 3 CO 2012 F3 8 RD 521 C1, E2 7 CO 2720 A1 RD 599 C1, C2, D2 D R CO 2720 B1 RD 647 H3 COBBS CV. -

Active with AAC
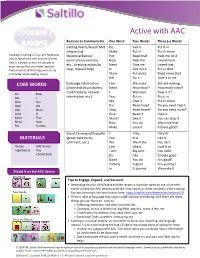
Active with AAC Reasons to Communicate One Word Two Words Three (+) Words This could be a great activity to use in many Getting Wants/Needs Met I I do I do it situations. You can play this at back-to- (requesting You I go You do it school time, with a new client, or anytime objects/activities/ My I help My turn to go you want to focus on social questions. You can change the questions up or you can permission/attention, Your You go Help me do it make more than one ball with different etc., directing action/to Turn Who go I will help stop, request help) Go Help me Who wants it questions. This could be a fun way to move away from the table while continuing to Help My turn Who wants a turn communicate. Who Help me Your turn to do it Exchange Information Who Who go Who is going next CORE WORDS (share and show objects, What Who turn Who gets it now confirm/deny, request Like Who want Who wants it Do Good Go I information, etc.) I What is What is your Help You You What your What is your favorite Like Your Mine What like I like XX Love Mine Go I like I love XX Will My Turn I love I love that too Favorite What Me My favorite I like it too Cool Who Too Me too My favorite is Fun That Love My turn It’s my turn Social Closeness/Etiquette I I go I want a go MATERIALS (greet, take turns, You Who go Who is going now comment, etc.) Mine My turn It’s my turn Beach ball Device My Your turn I like that Velcro/glue AAC user Turn I like I like it Questions You Like I love I love it (written on Love You like That’s really cool paper) Cool That’s cool I think that’s fun Good That’s good This is fun Model It on the AAC Device One Word: Tips to Engage, Expand, and Succeed: • To play: whenever someone catches the ball, whatever question is touching (or is closest to) their right thumb is the question they must answer. -

What Are Core Words Anyway? Let Us Help You Get Started! Part 1 Goals for AAC Users
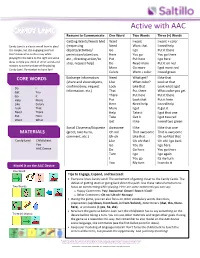
What Are Core Words Anyway? Let Us Help You Get Started! Part 1 Introductions and Presentation Goals Goals for AAC Users a. Learn cause and effect Reasons why we communicate b. Communicate with peers 1) Naming and adults c. Learn Language 2) Greeting d. Initiate – not just respond 3) Commenting e. Make friends 4) Requesting Objects f. Social skills g. Learn educational content 5) Protesting h. Participate in class or 6) Requesting Information workplace 7) Responding i. Gain independence – say 8) Requesting Action what they want to say! 9) Responding to Request 10) Regulating Conversational Behavior 11) Relaying Past Events Commenting 12) Stating Greeting Naming Activity to Build Communication Responding Requesting Objects Protesting Requesting Information Requesting Action Making a Snack Reading a Book Suggested Activity https://saltillo.com/ 1 © 2020 PRC-Saltillo. Non-commercial reprint rights for clinical or personal use granted with inclusion of copyright notice. Commercial use prohibited; may not be used for resale. Contact PRC-Saltillo for questions regarding permissible uses. Understand the target Various ways to look at language development, but ultimately they lead us down the same path • Developmental Milestones • Language acquisition stages • Browns Stages How can we aim for the target? Use CORE VOCABULARY! Notes Core vocabulary is: o Reusable across contexts, o Made up of approximately 250-400 words that account for 80% of our utterances, and is the same across gender, age, topic, setting, and disability. o Includes pronouns, verbs, adjectives, questions, interjections, prepositions and adverbs. Which class of words is missing? ______________ https://saltillo.com/ 2 © 2020 PRC-Saltillo. Non-commercial reprint rights for clinical or personal use granted with inclusion of copyright notice. -

AAC Basics and Implementation: How to Teach Students Who “Talk with Technology”
AAC Basics and Implementation: How to Teach Students who “Talk with Technology” Paul Visvader MA CCC-SLP Assistive Technology Team, Boulder Valley School District, Boulder, CO ©2013 AAC Basics and Implementation: How to Teach Students who “Talk with Technology” “Augmentative or Alternative Communication (AAC) is any device, system, or method that improves the ability of a child with a communication impairment to communicate effectively.” Paul Visvader MA CCC-SLP © 2013 Assistive Technology Team, Boulder Valley School District, Boulder, CO Table of Contents Introduction ..................................................................................................................................... 3 Background ...................................................................................................................................... 4 Learning communication skills .................................................................................................. 4 Three levels of communicative abilities .................................................................................... 5 Augmentative and alternative communication (AAC) .............................................................. 5 AAC Evaluation ................................................................................................................................. 7 The “AAC Toolbox” (a general discussion) ....................................................................................... 8 AAC Setup and Implementation ................................................................................................... -

Active with AAC
Active with AAC Reasons to Communicate One Word Two Words Three (+) Words Getting Wants/Needs Met Want I want I want that one Many children love to play board games! (requesting Need You need I need it But, you may be wondering what can I model and say while playing a board objects/activities/ That Put there Put it there game? There are plenty of core words you permission/attention, There Put on Put it on there can model while playing any board game etc., directing action/to Put Give that Give me that one you want! Check out the chart and our Tips stop, request help) Give Want that Help me open it to Engage, Expand, and Succeed! Help Help open Let’s play again Open Go + “color” Go to “color” Exchange Information Open Put on? I did it CORE WORDS (share and show objects, Like I do/did Put it there confirm/deny, request That You put Put in on here? Do I Give You information, etc.) There Need that I need that Go My Put What next? You need this one? Like Uh-oh On You win Open that one Lose Cool In I lose Let’s play more Need There Let’s Where put? I go next Put That Where Open that What is next? Want “Colors” What Let’s play I lose again Win Again Next I go I go again Social Closeness/Etiquette I My turn I did it MATERIALS (greet, take turns, You I go I go again comment, etc.) My You go Uh-oh! You lose Board Game You Turn I win Cool! I win! AAC User Go You lose Let’s play again AAC Device Win That cool That is “color” Lose Let’s play That was cool Uh-oh I do It’s my turn Colors That + “color” This is fun Model It on the AAC Device One Word: Tips to Engage, Expand, and Succeed: The most important thing to remember while you play is to keep it natural. -
Active with AAC
Active with AAC Reasons to Communicate One Word Two Words Three (+) Words Getting Wants/Needs Met Do Get it Put it in (requesting Make Put in Put in more Cooking, or baking, is a fun and functional objects/activities/ Put Need that Help me do it way to spend time with your child/client. permission/attention, Help Help me I need more And as a bonus, at the end, you get to enjoy eating what you made together! etc., directing action/to Need Give me I need that stop, request help) In Get more Put in that Take a look at all the things you can say and model while cooking. Enjoy! More Put more Need more that Get Do it Give it to me Exchange Information Like We make We are making CORE WORDS (share and show objects, Need How much? How much need? confirm/deny, request I We need How is it? Do Stop Get I information, etc.) You Put in Put it in Give You We Give it Put in more Help We Put Need help? Do you need help? Like More Stop Need more? Do you need more? Look In Give Need it I like it Make That More? Like it You can stop it Need How How You do We make that Put Yummy Make Look it It looks good! Social Closeness/Etiquette I I like I like it! MATERIALS (greet, take turns, You It in I do it comment, etc.) We We make You do it Recipe AAC Device Like Like it I put it in Ingredients You Turn My turn You did it Child/Client Do I do It looks good Good You do It is good! Yummy It good It is yummy! In It yummy We make it Model It on the AAC Device One Word: Tips to Engage, Expand, and Succeed: • Depending on your child/client and the recipe, it might be safest to use a low-tech board to keep the AAC device protected from liquids or other accidents while cooking. -

User's Guide for Dedicated Model
User’s Guide for Dedicated Model Saltillo Corporation 2143 Township Road 112 Millersburg, OH 44654 www.saltillo.com NOVA chat and VocabPC are trademarks of Saltillo Corporation. Microsoft and Windows are either registered trademarks or trademarks of Microsoft Corporation in the United States and/or other countries. Android is a trademark of Google Inc. Samsung and Galaxy Tab are registered trademarks of Samsung Electronics America, Inc. Android Market is a trademark of Google Inc. Neospeech is a trademark of Neospeech Inc. Loquendo is a trademark of Loquendo S.p.A. Ivona is Copyright © 2001-2010, IVO Software Sp.z o.o. Acapela is a trademark of Acapela Group PCS Symbols illustrations are copyright of Mayer-Johnson Co. Symbol Stix is copyright of News2You. "The Library of Character/Logo Symbols contained in this software is included free of charge, may be used solely for communication purposes and may not be sold, copied or otherwise exploited for any type of profit." WordPower and ChatPower is copyright of Inman Innovations My QuickChat 12 Adult is copyright of Talk To Me Technologies My QuickChat 12 Child is copyright of Talk To Me Technologies My QuickChat 8 Adult is copyright of Talk To Me Technologies My QuickChat 8 Child is copyright of Talk To Me Technologies My QuickChat 4 Child is copyright of Talk To Me Technologies NOVA chat Editor CD NOVA chat Editor, owned by Saltillo Corporation Microsoft Voices, owned by Microsoft Corporation Microsoft's agreement states: INSTALLATION AND USE RIGHTS. You may install and use one copy of the software on each computer on your premises that you use to exchange data and software with portable devices powered by a Microsoft operating system. -

School-Based Aac Evaluation
SCHOOL -BASED AAC EVALUATION: Choosing Effective Assistive Technology Strategies for Students with Complex Communication Needs Paul Visvader, MA CCC-SLP SCHOOL-BASED AAC EVALUATION: Choosing Effective Assistive Technology Strategies for Students with Complex Communication Needs AAC: “Augmentative or Alternative Communication (AAC) is any device, system, or method that improves the ability of a child with a communication impairment to communicate effectively.” By Paul Visvader MA CCC-SLP <[email protected]> Boulder, Colorado “Share the aloha…” Dedicated to all of my A.T. team members, past, present and future (Margaret Janda, Georgia Magnera, Rosemary Bogart, Anja Kintsch, Nancy Lamb, Brittany Horlbeck, Jennifer Leonesio, Erika Brandstatter...); to colleagues, friends, and mentors; and of course, to all of our students. (The original version of this book was posted online as “School-Based Assistive Technology Communication Evaluation” ©2011—It is revised and published here with permission) Copyright ©2015 by Paul Visvader <[email protected]> All rights reserved. Table of Contents Introduction ..................................................................................................................................... 1 Chapter 1: Background .................................................................................................................... 3 Learning communication skills .................................................................................................. 3 Three levels of communicative abilities ................................................................................... -
Active with AAC
Active with AAC Reasons to Communicate One Word Two Words Three (+) Words Getting Wants/Needs Met Want I want I want + color Candy Land is a classic we all love to play! (requesting Need Want that I need help It is simple, but still engaging and fun! objects/activities/ Go I go Put it there Don’t know what to do or say while permission/attention, Here You go You go there playing? In the table to the right are some etc., directing action/to Put Put here I go here ideas to help you think of other words and stop, request help) Do Need more Put it on red reasons to communicate while playing More Go more I got more red Candy Land. Remember to have fun! Colors Want + color I need green Exchange Information Need What get? I like that CORE WORDS (share and show objects, Like What color? Look at that confirm/deny, request Look Like that Look what I got Do I Get You information, etc.) That Put there What color you get Go It There Put here Put it there Help More Put Look that Put it here Like Colors Here Need help I need help Look That More I got It got it Need There Help Take it I got that one Put Here Take Get it I got two red Want What Get I like I need two green Social Closeness/Etiquette Awesome I like I like that one MATERIALS (greet, take turns, Oh no! That awesome That is awesome comment, etc.) Uh-oh Like that Oh no! Not that Candy Land Child/client Like Uh-oh that! Uh-oh! I go back You Go You do I go here AAC Device Do Go here You go there Turn I go I go again I You go Its my turn You My turn I can do it Model It on the AAC Device One Word: Tips to Engage, Expand, and Succeed: Everyone loves Candy Land! The excitement of getting closer to the Candy Castle. -

C-1 Charter of the Town of Saltillo, Tennessee1
C-1 CHARTER OF THE TOWN OF SALTILLO, TENNESSEE1 CHAPTER NO. 45 HOUSE BILL NO. 2391 By Representative Rinks Substituted for: Senate Bill No. 2379 By Mr. Speaker Wilder AN ACT to amend Chapter 699 of the Private Acts of 1951; as amended by Chapter 225 of the Private Acts of 1976; Chapter 3 of the Private Acts of 1991 and Chapter 133 of the Private Acts of 2004; and any other acts amendatory thereto, relative to the charter for the Town of Saltillo. TABLE OF CONTENTS SECTION PAGE (1) Repeals Priv. Acts 1951, Ch. 699 except for boundaries .......... C-2 (2) Incorporation ........................................... C-2 (3) Definitions ............................................. C-2 (4) Boundaries ............................................. C-3 (5) Board of mayor and aldermen to be legislative power; term of office ................................................ C-3 (6) Elections; voter qualifications; contests of election ............. C-3 (7) Mayor and aldermen--oath of office.......................... C-4 (8) Vacancies in board....................................... C-4 (9) Meetings of board of mayor and aldermen .................... C-4 (10) Board to constitute quorum; recorder to keep minutes .......... C-4 (11) Ordinance powers of the board ............................. C-5 1Priv. Acts 2005, ch. 45, is the current basic charter act for the Town of Saltillo, Tennessee. The text of the basic charter act set out herein includes all its amendments through the 2020 session of the Tennessee General Assembly. Sections of the charter which have been amended contain at the end of those sections the citation to the official private act or acts constituting the amendment or amendments. No other changes have been made to the charter except the addition of a table of contents to facilitate its use. -

Augmentative and Alternative Communication Assessment
Disclosures Let’s Get This Started! Augmentative and Alternative Financial Disclosures: YES Communication Assessment Non- Financial Disclosures: YES Presented by: Heather Miller Schwarz, M.S., CCC-SLP, UCP Seguin- Infinitec © H. Miller-Schwarz, 2019 1 2 Sometimes you learn what is “right” by being “wrong”. © H. Miller-Schwarz, 2019 © H. Miller-Schwarz, 2019 3 4 Goals for this Training • Participants will compare and contrast models of AAC assessment and strategy implementation. • Participants will identify potential AAC users using the participation model as a guide. • Participants will list the components of the feature match assessment to support communication. © H. Miller-Schwarz, 2019 © H. Miller-Schwarz, 2018 5 6 1 AAC Assessment AAC Assessment The goals of AAC intervention and assessment • • Which communication methods should be • Enable individuals to efficiently and effectively maintained or developed? engage in communication and participate in • Which communication tools and methods will activities of their choice. be added to the student’s repertoire? • Assist individuals to meet their current and future communication needs. • What strategies will the communication partners use with this individualto increase communicative competence? http://aac.unl.edu/yaack/c0.html © K. Post 7 H. Miller-Schwarz, 2018 7 8 AAC Assessment Assessment Models family person with CCN • Expert Model A team approach. teachers/staff • Formal referral system • Formal assessment • Decision made with the help of an ‘expert’ caregivers doctors • Formal report • Then, implementation shifts to team working with this individual therapists • This team is responsible for implementation and determining success of a system close friends © K. Post 7 H. Miller-Schwarz, 2018 Bowser and Decoste, 2016 © H.