Changes of Gut-Microbiota-Liver Axis in Hepatitis C Virus Infection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prevention & Control of Viral Hepatitis Infection
Prevention & Control of Viral Hepatitis Infection: A Strategy for Global Action © World Health Organization 2011. All rights reserved. The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by WHO to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either express or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. Table of contents Disease burden 02 What is viral hepatitis? 05 Prevention & control: a tailored approach 06 Global Achievements 08 Remaining challenges 10 World Health Assembly: a mandate for comprehensive prevention & control 13 WHO goals and strategy -

The Role of Hepatitis C Virus in Hepatocellular Carcinoma U
Viruses in cancer cell plasticity: the role of hepatitis C virus in hepatocellular carcinoma U. Hibner, D. Gregoire To cite this version: U. Hibner, D. Gregoire. Viruses in cancer cell plasticity: the role of hepatitis C virus in hepato- cellular carcinoma. Contemporary Oncology, Termedia Publishing House, 2015, 19 (1A), pp.A62–7. 10.5114/wo.2014.47132. hal-02187396 HAL Id: hal-02187396 https://hal.archives-ouvertes.fr/hal-02187396 Submitted on 2 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License Review Viruses are considered as causative agents of a significant proportion of human cancers. While the very Viruses in cancer cell plasticity: stringent criteria used for their clas- sification probably lead to an under- estimation, only six human viruses the role of hepatitis C virus are currently classified as oncogenic. In this review we give a brief histor- in hepatocellular carcinoma ical account of the discovery of on- cogenic viruses and then analyse the mechanisms underlying the infectious causes of cancer. We discuss viral strategies that evolved to ensure vi- Urszula Hibner1,2,3, Damien Grégoire1,2,3 rus propagation and spread can alter cellular homeostasis in a way that increases the probability of oncogen- 1Institut de Génétique Moléculaire de Montpellier, CNRS, UMR 5535, Montpellier, France ic transformation and acquisition of 2Université Montpellier 2, Montpellier, France stem cell phenotype. -

Rational Engineering of HCV Vaccines for Humoral Immunity
viruses Review To Include or Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity Felicia Schlotthauer 1,2,†, Joey McGregor 1,2,† and Heidi E Drummer 1,2,3,* 1 Viral Entry and Vaccines Group, Burnet Institute, Melbourne, VIC 3004, Australia; [email protected] (F.S.); [email protected] (J.M.) 2 Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, VIC 3000, Australia 3 Department of Microbiology, Monash University, Clayton, VIC 3800, Australia * Correspondence: [email protected]; Tel.: +61-392-822-179 † These authors contributed equally to this work. Abstract: Direct-acting antiviral agents have proven highly effective at treating existing hepatitis C infections but despite their availability most countries will not reach the World Health Organization targets for elimination of HCV by 2030. A prophylactic vaccine remains a high priority. Whilst early vaccines focused largely on generating T cell immunity, attention is now aimed at vaccines that gen- erate humoral immunity, either alone or in combination with T cell-based vaccines. High-resolution structures of hepatitis C viral glycoproteins and their interaction with monoclonal antibodies isolated from both cleared and chronically infected people, together with advances in vaccine technologies, provide new avenues for vaccine development. Keywords: glycoprotein E2; vaccine development; humoral immunity Citation: Schlotthauer, F.; McGregor, J.; Drummer, H.E To Include or 1. Introduction Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity. The development of direct-acting antiviral agents (DAA) with their ability to cure Viruses 2021, 13, 805. https:// infection in >95% of those treated was heralded as the key to eliminating hepatitis C globally. -
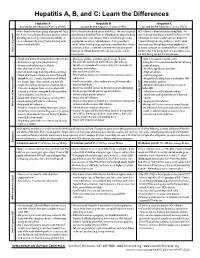
Hepatitis A, B, and C: Learn the Differences
Hepatitis A, B, and C: Learn the Differences Hepatitis A Hepatitis B Hepatitis C caused by the hepatitis A virus (HAV) caused by the hepatitis B virus (HBV) caused by the hepatitis C virus (HCV) HAV is found in the feces (poop) of people with hepa- HBV is found in blood and certain body fluids. The virus is spread HCV is found in blood and certain body fluids. The titis A and is usually spread by close personal contact when blood or body fluid from an infected person enters the body virus is spread when blood or body fluid from an HCV- (including sex or living in the same household). It of a person who is not immune. HBV is spread through having infected person enters another person’s body. HCV can also be spread by eating food or drinking water unprotected sex with an infected person, sharing needles or is spread through sharing needles or “works” when contaminated with HAV. “works” when shooting drugs, exposure to needlesticks or sharps shooting drugs, through exposure to needlesticks on the job, or from an infected mother to her baby during birth. or sharps on the job, or sometimes from an infected How is it spread? Exposure to infected blood in ANY situation can be a risk for mother to her baby during birth. It is possible to trans- transmission. mit HCV during sex, but it is not common. • People who wish to be protected from HAV infection • All infants, children, and teens ages 0 through 18 years There is no vaccine to prevent HCV. -

Understanding Human Astrovirus from Pathogenesis to Treatment
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 6-2020 Understanding Human Astrovirus from Pathogenesis to Treatment Virginia Hargest University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Diseases Commons, Medical Sciences Commons, and the Viruses Commons Recommended Citation Hargest, Virginia (0000-0003-3883-1232), "Understanding Human Astrovirus from Pathogenesis to Treatment" (2020). Theses and Dissertations (ETD). Paper 523. http://dx.doi.org/10.21007/ etd.cghs.2020.0507. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Understanding Human Astrovirus from Pathogenesis to Treatment Abstract While human astroviruses (HAstV) were discovered nearly 45 years ago, these small positive-sense RNA viruses remain critically understudied. These studies provide fundamental new research on astrovirus pathogenesis and disruption of the gut epithelium by induction of epithelial-mesenchymal transition (EMT) following astrovirus infection. Here we characterize HAstV-induced EMT as an upregulation of SNAI1 and VIM with a down regulation of CDH1 and OCLN, loss of cell-cell junctions most notably at 18 hours post-infection (hpi), and loss of cellular polarity by 24 hpi. While active transforming growth factor- (TGF-) increases during HAstV infection, inhibition of TGF- signaling does not hinder EMT induction. However, HAstV-induced EMT does require active viral replication. -

Metagenomic Identification of Diverse Animal Hepaciviruses and Pegiviruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.16.100149; this version posted May 17, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Metagenomic identification of diverse animal hepaciviruses and 2 pegiviruses 3 4 5 Ashleigh F. Porter1, John H.-O. Pettersson1,2, Wei-Shan Chang1, Erin Harvey1, Karrie Rose3, 6 Mang Shi5, John-Sebastian Eden1,4, Jan Buchmann1, Craig Moritz6, Edward C. Holmes1 7 8 9 1. Marie Bashir Institute for Infectious Diseases and Biosecurity, School of Life and 10 Environmental Sciences and School of Medical Sciences, The University of Sydney, 11 Sydney, Australia. 12 2. Zoonosis Science Center, Department of Medical Biochemistry and Microbiology, 13 Uppsala University, Uppsala, Sweden. 14 3. Australian Registry of Wildlife Health, Taronga Conservation Society Australia, 15 Mosman, Australia. 16 4. Centre for Virus Research, Westmead Institute for Medical Research, Westmead, 17 Australia. 18 5. School of Medicine, Sun Yat-sen University, Guangzhou, China. 19 6. Research School of Biology, and Centre for Biodiversity Analysis, Australian National 20 University, Acton, ACT, Australia. 21 22 23 Address correspondence to: 24 Edward C. Holmes, 25 Marie Bashir Institute for Infectious Diseases and Biosecurity, School of Life and 26 Environmental Sciences and School of Medical Sciences, The University of Sydney, 27 Sydney, Australia 28 [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.16.100149; this version posted May 17, 2020. -

Host Cell Functions in Vesicular Stomatitis Virus Replication Phat X
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Veterinary and Biomedical Science Veterinary and Biomedical Sciences, Department of 2016 Host Cell Functions In Vesicular Stomatitis Virus Replication Phat X. Dinh University of Nebraska - Lincoln Anshuman Das University of Nebraska-Lincoln Asit K. Pattnaik University of Nebraska - Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/vetscipapers Part of the Biochemistry, Biophysics, and Structural Biology Commons, Cell and Developmental Biology Commons, Immunology and Infectious Disease Commons, Medical Sciences Commons, Veterinary Microbiology and Immunobiology Commons, and the Veterinary Pathology and Pathobiology Commons Dinh, Phat X.; Das, Anshuman; and Pattnaik, Asit K., "Host Cell Functions In Vesicular Stomatitis Virus Replication" (2016). Papers in Veterinary and Biomedical Science. 237. https://digitalcommons.unl.edu/vetscipapers/237 This Article is brought to you for free and open access by the Veterinary and Biomedical Sciences, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Papers in Veterinary and Biomedical Science by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published in Biology and Pathogenesis of Rhabdo and Filoviruses Copyright © 2016 World Scientific Publishing Co Pte Ltd digitalcommons.unl.edu HOST CELL FUNCTIONS IN VESICULAR STOMATITIS VIRUS REPLICATION Phat X. Dinh, Anshuman Das, and Asit K. Pattnaik School of Veterinary Medicine and Biomedical Sciences and the Nebraska Center for Virology, University of Nebraska-Lincoln, 109 Morrison Life Science Research Center, 4240 Fair Street, Lincoln, * Nebraska 68583, USA E-mail: [email protected] Summary Vesicular stomatitis virus (VSV), the prototypic rhabdovirus, has been used as an excellent paradigm for understanding the mechanisms of virus replication, pathogenesis, host response to virus infection and also for myriads of studies on cellular and molecular biology. -

HCV Eradication with Direct Acting Antivirals (Daas)?
HCV eradication with direct acting antivirals (DAAs)? Emilie Estrabaud Service d’Hépatologie et INSERM UMR1149, AP-HP Hôpital Beaujon, Paris, France. [email protected] HCV eradication with direct acting antivirals (DAAs)? HCV replication HCV genome and DAAs targets NS3 inhibitors NS5A inhibitors NS5B inhibitors Take home messages HCV viral cycle Asselah et al. Liver Int. 2015;35 S1:56-64. Direct acting antivirals 5’NTR Structural proteins Nonstructural proteins 3’NTR Metalloprotease Envelope Serine protease Glycoproteins RNA Capsid RNA helicase Cofactors Polymerase C E1 E2 NS1 NS2 NS3 NS4A NS4B NS5A NS5B Protease Inhibitors NS5A Inhibitors Polymerase Inhibitors Telaprevir Daclatasvir Nucs Non-Nucs Boceprevir Ledipasvir Simeprevir ABT-267 Sofosbuvir ABT-333 Faldaprevir GS-5816 VX-135 Deleobuvir Asunaprevir Direct Acting Antivirals: 2015 Asselah et al. Liver Int. 2015;35 S1:56-64. Genetic barrier for HCV direct acting antivirals High Nucleos(t)ide 1 mutation= high cost to Analog Inhibitors fitness, 2-3 additional mutations to increase fitness 2 st generation Protease Inhibitors n Non Nucleos(t)ide Analog Inhibitors : NS5 A Inhibitors 1 st generation Protease Inhibitors 1 mutation= low cost to fitness Low Halfon et al. J Hepatol 2011. Vol 55(1):192-206. HCV protease inhibitors (PI) Inhibit NS3/NS4A serine protease responsible for the processing of the polyprotein 1st generation 1st generation, 2nd wave 2nd generation Resistance low low high barrier Genotype activity 1: 1 a< 1b All except 3 all Drug drug Important Less Less interaction Drugs Boceprevir Simeprevir (Janssen) MK-5172 Telaprevir Faldaprevir (BI) ACH-2684 Paritaprevir (ABT-450)/r (AbbVie) Vedroprevir (Gilead) Vaniprevir (Merck) Sovaprevir (Achillion) Asunaprevir (BMS) NS3/NS4A structure Repositioning of helicase domain Self cleavage Lipid Bilayer Inactive Insertion of the Active carboxy-terminal tail Bartenschlager et al. -

Origins and Evolution of the Global RNA Virome
bioRxiv preprint doi: https://doi.org/10.1101/451740; this version posted October 24, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Origins and Evolution of the Global RNA Virome 2 Yuri I. Wolfa, Darius Kazlauskasb,c, Jaime Iranzoa, Adriana Lucía-Sanza,d, Jens H. 3 Kuhne, Mart Krupovicc, Valerian V. Doljaf,#, Eugene V. Koonina 4 aNational Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland, USA 5 b Vilniaus universitetas biotechnologijos institutas, Vilnius, Lithuania 6 c Département de Microbiologie, Institut Pasteur, Paris, France 7 dCentro Nacional de Biotecnología, Madrid, Spain 8 eIntegrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious 9 Diseases, National Institutes of Health, Frederick, Maryland, USA 10 fDepartment of Botany and Plant Pathology, Oregon State University, Corvallis, Oregon, USA 11 12 #Address correspondence to Valerian V. Dolja, [email protected] 13 14 Running title: Global RNA Virome 15 16 KEYWORDS 17 virus evolution, RNA virome, RNA-dependent RNA polymerase, phylogenomics, horizontal 18 virus transfer, virus classification, virus taxonomy 1 bioRxiv preprint doi: https://doi.org/10.1101/451740; this version posted October 24, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 19 ABSTRACT 20 Viruses with RNA genomes dominate the eukaryotic virome, reaching enormous diversity in 21 animals and plants. The recent advances of metaviromics prompted us to perform a detailed 22 phylogenomic reconstruction of the evolution of the dramatically expanded global RNA virome. -

Caracterización Molecular Del Perfil De Resistencias Del Virus De La
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Programa de doctorado en Medicina Departamento de Medicina Facultad de Medicina Universidad Autónoma de Barcelona TESIS DOCTORAL Caracterización molecular del perfil de resistencias del virus de la hepatitis C después del fallo terapéutico a antivirales de acción directa mediante secuenciación masiva Tesis para optar al grado de doctor de Qian Chen Directores de la Tesis Dr. Josep Quer Sivila Dra. Celia Perales Viejo Dr. Josep Gregori i Font Laboratorio de Enfermedades Hepáticas - Hepatitis Víricas Vall d’Hebron Institut de Recerca (VHIR) Barcelona, 2018 ABREVIACIONES Abreviaciones ADN: Ácido desoxirribonucleico AK: Adenosina quinasa ALT: Alanina aminotransferasa ARN: Ácido ribonucleico ASV: Asunaprevir BOC: Boceprevir CCD: Charge Coupled Device CLDN1: Claudina-1 CHC: Carcinoma hepatocelular DAA: Antiviral de acción directa DC-SIGN: Dendritic cell-specific ICAM-3 grabbing non-integrin DCV: Daclatasvir DSV: Dasabuvir -

REVIEW Hepatitis C Virus
DOI:http://dx.doi.org/10.7314/APJCP.2012.13.12.5917 Hepatitis C Virus -Proteins, Diagnosis, Treatment and New Approaches for Vaccine Development REVIEW Hepatitis C Virus - Proteins, Diagnosis, Treatment and New Approaches for Vaccine Development Hossein Keyvani1, Mehdi Fazlalipour2*, Seyed Hamid Reza Monavari1, Hamid Reza Mollaie2 Abstract Background: Hepatitis C virus (HCV) causes acute and chronic human hepatitis infection and as such is an important global health problem. The virus was discovered in the USA in 1989 and it is now known that three to four million people are infected every year, WHO estimating that 3 percent of the 7 billion people worldwide being chronically infected. Humans are the natural hosts of HCV and this virus can eventually lead to permanent liver damage and carcinoma. HCV is a member of the Flaviviridae family and Hepacivirus genus. The diameter of the virus is about 50-60 nm and the virion contains a single-stranded positive RNA approximately 10,000 nucleotides in length and consisting of one ORF which is encapsulated by an external lipid envelope and icosahedral capsid. HCV is a heterogeneous virus, classified into 6 genotypes and more than 50 subtypes. Because of the genome variability, nucleotide sequences of genotypes differ by approximately 31-34%, and by 20-23% among subtypes. Quasi-species of mixed virus populations provide a survival advantage for the virus to create multiple variant genomes and a high rate of generation of variants to allow rapid selection of mutants for new environmental conditions. Direct contact with infected blood and blood products, sexual relationships and availability of injectable drugs have had remarkable effects on HCV epidemiology. -

Arenaviridae Astroviridae Filoviridae Flaviviridae Hantaviridae
Hantaviridae 0.7 Filoviridae 0.6 Picornaviridae 0.3 Wenling red spikefish hantavirus Rhinovirus C Ahab virus * Possum enterovirus * Aronnax virus * * Wenling minipizza batfish hantavirus Wenling filefish filovirus Norway rat hunnivirus * Wenling yellow goosefish hantavirus Starbuck virus * * Porcine teschovirus European mole nova virus Human Marburg marburgvirus Mosavirus Asturias virus * * * Tortoise picornavirus Egyptian fruit bat Marburg marburgvirus Banded bullfrog picornavirus * Spanish mole uluguru virus Human Sudan ebolavirus * Black spectacled toad picornavirus * Kilimanjaro virus * * * Crab-eating macaque reston ebolavirus Equine rhinitis A virus Imjin virus * Foot and mouth disease virus Dode virus * Angolan free-tailed bat bombali ebolavirus * * Human cosavirus E Seoul orthohantavirus Little free-tailed bat bombali ebolavirus * African bat icavirus A Tigray hantavirus Human Zaire ebolavirus * Saffold virus * Human choclo virus *Little collared fruit bat ebolavirus Peleg virus * Eastern red scorpionfish picornavirus * Reed vole hantavirus Human bundibugyo ebolavirus * * Isla vista hantavirus * Seal picornavirus Human Tai forest ebolavirus Chicken orivirus Paramyxoviridae 0.4 * Duck picornavirus Hepadnaviridae 0.4 Bildad virus Ned virus Tiger rockfish hepatitis B virus Western African lungfish picornavirus * Pacific spadenose shark paramyxovirus * European eel hepatitis B virus Bluegill picornavirus Nemo virus * Carp picornavirus * African cichlid hepatitis B virus Triplecross lizardfish paramyxovirus * * Fathead minnow picornavirus