A Novel Method to Rescue and Culture Duck Astrovirus Type 1 in Vitro
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Viral Hepatitis Testing Effective Date: January 1, 2012
Viral Hepatitis Testing Effective Date: January 1, 2012 Scope This guideline provides guidance for the use of laboratory tests to diagnose acute and chronic viral hepatitis in adults (> 19 years) in the primary care setting. General Considerations for Ordering Laboratory Tests Prior to ordering tests for hepatitis, consider the patient’s history, age, risk factors (see below), hepatitis vaccination status, and any available previous hepatitis test results. Risk Factors for Viral Hepatitis include: • Substance use (includes sharing drug snorting, smoking or injection equipment) • High-risk sexual activity or sexual partner with viral hepatitis • Travel to or from high-risk hepatitis endemic areas or exposure during a local outbreak • Immigration from hepatitis B and/or C endemic countries • Household contact with an infected person especially if personal items (e.g., razors, toothbrushes, nail clippers) are shared • Recipient of unscreened blood products* • Needle-stick injury or other occupational exposure (e.g., healthcare workers) • Children born to mothers with chronic hepatitis B or C infection • Attendance at daycare • Contaminated food or water (hepatitis A only) • Tattoos and body piercing • History of incarceration • HIV or other sexually transmitted infection • Hemodialysis *screening of donated blood products for hepatitis C (anti-HCV) began in 1990 in Canada.1 Types of Viral Hepatitis Hepatitis A: causes acute but not chronic hepatitis Hepatitis B: causes acute and chronic hepatitis Hepatitis C: causes chronic hepatitis but rarely manifests as acute hepatitis Hepatitis D: rare and only occurs in patients infected with hepatitis B Hepatitis E: clinically similar to hepatitis A, mostly restricted to endemic areas and occasionally causes chronic infection in immunosuppressed people Others: e.g. -

Prevention & Control of Viral Hepatitis Infection
Prevention & Control of Viral Hepatitis Infection: A Strategy for Global Action © World Health Organization 2011. All rights reserved. The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by WHO to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either express or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. Table of contents Disease burden 02 What is viral hepatitis? 05 Prevention & control: a tailored approach 06 Global Achievements 08 Remaining challenges 10 World Health Assembly: a mandate for comprehensive prevention & control 13 WHO goals and strategy -

The Role of Hepatitis C Virus in Hepatocellular Carcinoma U
Viruses in cancer cell plasticity: the role of hepatitis C virus in hepatocellular carcinoma U. Hibner, D. Gregoire To cite this version: U. Hibner, D. Gregoire. Viruses in cancer cell plasticity: the role of hepatitis C virus in hepato- cellular carcinoma. Contemporary Oncology, Termedia Publishing House, 2015, 19 (1A), pp.A62–7. 10.5114/wo.2014.47132. hal-02187396 HAL Id: hal-02187396 https://hal.archives-ouvertes.fr/hal-02187396 Submitted on 2 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License Review Viruses are considered as causative agents of a significant proportion of human cancers. While the very Viruses in cancer cell plasticity: stringent criteria used for their clas- sification probably lead to an under- estimation, only six human viruses the role of hepatitis C virus are currently classified as oncogenic. In this review we give a brief histor- in hepatocellular carcinoma ical account of the discovery of on- cogenic viruses and then analyse the mechanisms underlying the infectious causes of cancer. We discuss viral strategies that evolved to ensure vi- Urszula Hibner1,2,3, Damien Grégoire1,2,3 rus propagation and spread can alter cellular homeostasis in a way that increases the probability of oncogen- 1Institut de Génétique Moléculaire de Montpellier, CNRS, UMR 5535, Montpellier, France ic transformation and acquisition of 2Université Montpellier 2, Montpellier, France stem cell phenotype. -

Rational Engineering of HCV Vaccines for Humoral Immunity
viruses Review To Include or Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity Felicia Schlotthauer 1,2,†, Joey McGregor 1,2,† and Heidi E Drummer 1,2,3,* 1 Viral Entry and Vaccines Group, Burnet Institute, Melbourne, VIC 3004, Australia; [email protected] (F.S.); [email protected] (J.M.) 2 Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, VIC 3000, Australia 3 Department of Microbiology, Monash University, Clayton, VIC 3800, Australia * Correspondence: [email protected]; Tel.: +61-392-822-179 † These authors contributed equally to this work. Abstract: Direct-acting antiviral agents have proven highly effective at treating existing hepatitis C infections but despite their availability most countries will not reach the World Health Organization targets for elimination of HCV by 2030. A prophylactic vaccine remains a high priority. Whilst early vaccines focused largely on generating T cell immunity, attention is now aimed at vaccines that gen- erate humoral immunity, either alone or in combination with T cell-based vaccines. High-resolution structures of hepatitis C viral glycoproteins and their interaction with monoclonal antibodies isolated from both cleared and chronically infected people, together with advances in vaccine technologies, provide new avenues for vaccine development. Keywords: glycoprotein E2; vaccine development; humoral immunity Citation: Schlotthauer, F.; McGregor, J.; Drummer, H.E To Include or 1. Introduction Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity. The development of direct-acting antiviral agents (DAA) with their ability to cure Viruses 2021, 13, 805. https:// infection in >95% of those treated was heralded as the key to eliminating hepatitis C globally. -
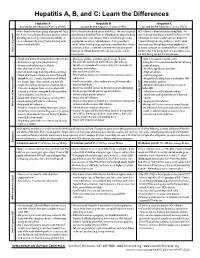
Hepatitis A, B, and C: Learn the Differences
Hepatitis A, B, and C: Learn the Differences Hepatitis A Hepatitis B Hepatitis C caused by the hepatitis A virus (HAV) caused by the hepatitis B virus (HBV) caused by the hepatitis C virus (HCV) HAV is found in the feces (poop) of people with hepa- HBV is found in blood and certain body fluids. The virus is spread HCV is found in blood and certain body fluids. The titis A and is usually spread by close personal contact when blood or body fluid from an infected person enters the body virus is spread when blood or body fluid from an HCV- (including sex or living in the same household). It of a person who is not immune. HBV is spread through having infected person enters another person’s body. HCV can also be spread by eating food or drinking water unprotected sex with an infected person, sharing needles or is spread through sharing needles or “works” when contaminated with HAV. “works” when shooting drugs, exposure to needlesticks or sharps shooting drugs, through exposure to needlesticks on the job, or from an infected mother to her baby during birth. or sharps on the job, or sometimes from an infected How is it spread? Exposure to infected blood in ANY situation can be a risk for mother to her baby during birth. It is possible to trans- transmission. mit HCV during sex, but it is not common. • People who wish to be protected from HAV infection • All infants, children, and teens ages 0 through 18 years There is no vaccine to prevent HCV. -

Active Peptic Ulcer Disease in Patients with Hepatitis C Virus-Related Cirrhosis: the Role of Helicobacter Pylori Infection and Portal Hypertensive Gastropathy
dore.qxd 7/19/2004 11:24 AM Page 521 View metadata, citation and similar papers at core.ac.uk ORIGINAL ARTICLE brought to you by CORE provided by Crossref Active peptic ulcer disease in patients with hepatitis C virus-related cirrhosis: The role of Helicobacter pylori infection and portal hypertensive gastropathy Maria Pina Dore MD PhD, Daniela Mura MD, Stefania Deledda MD, Emmanouil Maragkoudakis MD, Antonella Pironti MD, Giuseppe Realdi MD MP Dore, D Mura, S Deledda, E Maragkoudakis, Ulcère gastroduodénal évolutif chez les A Pironti, G Realdi. Active peptic ulcer disease in patients patients atteints de cirrhose liée au HCV : Le with hepatitis C virus-related cirrhosis: The role of Helicobacter pylori infection and portal hypertensive rôle de l’infection à Helicobacter pylori et de la gastropathy. Can J Gastroenterol 2004;18(8):521-524. gastropathie liée à l’hypertension portale BACKGROUND & AIM: The relationship between Helicobacter HISTORIQUE ET BUT : Le lien entre l’infection à Helicobacter pylori pylori infection and peptic ulcer disease in cirrhosis remains contro- et l’ulcère gastroduodénal dans la cirrhose reste controversé. Le but de la versial. The purpose of the present study was to investigate the role of présente étude est de vérifier le rôle de l’infection à H. pylori et de la gas- H pylori infection and portal hypertension gastropathy in the preva- tropathie liée à l’hypertension portale dans la prévalence de l’ulcère gas- lence of active peptic ulcer among dyspeptic patients with compen- troduodénal évolutif chez les patients dyspeptiques souffrant d’une sated hepatitis C virus (HCV)-related cirrhosis. -

Understanding Human Astrovirus from Pathogenesis to Treatment
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 6-2020 Understanding Human Astrovirus from Pathogenesis to Treatment Virginia Hargest University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Diseases Commons, Medical Sciences Commons, and the Viruses Commons Recommended Citation Hargest, Virginia (0000-0003-3883-1232), "Understanding Human Astrovirus from Pathogenesis to Treatment" (2020). Theses and Dissertations (ETD). Paper 523. http://dx.doi.org/10.21007/ etd.cghs.2020.0507. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Understanding Human Astrovirus from Pathogenesis to Treatment Abstract While human astroviruses (HAstV) were discovered nearly 45 years ago, these small positive-sense RNA viruses remain critically understudied. These studies provide fundamental new research on astrovirus pathogenesis and disruption of the gut epithelium by induction of epithelial-mesenchymal transition (EMT) following astrovirus infection. Here we characterize HAstV-induced EMT as an upregulation of SNAI1 and VIM with a down regulation of CDH1 and OCLN, loss of cell-cell junctions most notably at 18 hours post-infection (hpi), and loss of cellular polarity by 24 hpi. While active transforming growth factor- (TGF-) increases during HAstV infection, inhibition of TGF- signaling does not hinder EMT induction. However, HAstV-induced EMT does require active viral replication. -

Astrovirus Evolution and Emergence T ⁎ Nicholas Wohlgemutha, Rebekah Honcea,B, Stacey Schultz-Cherrya, a Department of Infectious Diseases, St
Infection, Genetics and Evolution 69 (2019) 30–37 Contents lists available at ScienceDirect Infection, Genetics and Evolution journal homepage: www.elsevier.com/locate/meegid Conference report Astrovirus evolution and emergence T ⁎ Nicholas Wohlgemutha, Rebekah Honcea,b, Stacey Schultz-Cherrya, a Department of Infectious Diseases, St. Jude Children's Research Hospital, Memphis, TN 38105, United States b Department of Microbiology, Immunology, and Biochemistry, University of Tennessee Health Science Center, Memphis, TN 38105, United States ARTICLE INFO ABSTRACT Keywords: Astroviruses are small, non-enveloped, positive-sense, single-stranded RNA viruses that belong to the Astroviridae Astrovirus family. Astroviruses infect diverse hosts and are typically associated with gastrointestinal illness; although Cross-species transmission disease can range from asymptomatic to encephalitis depending on the host and viral genotype. Astroviruses Recombination have high genetic variability due to an error prone polymerase and frequent recombination events between Emergence strains. Once thought to be species specific, recent evidence suggests astroviruses can spread between different host species, although the frequency with which this occurs and the restrictions that regulate the process are unknown. Recombination events can lead to drastic evolutionary changes and contribute to cross-species transmission events. This work reviews the current state of research on astrovirus evolution and emergence, especially as it relates to cross-species transmission and recombination of astroviruses. 1. Introduction 2. Genetics and genomics Astroviruses are nonenveloped, show icosahedral morphology, and Little is known about the astrovirus genome compared to other, have positive-sense, single-stranded RNA (+ssRNA) genomes (Méndez better characterized viruses. While several virus replication processes and Arias, 2013). They infect a multitude of hosts from birds to mam- and structural elements have been mapped to the genome (Fig. -

Hepatitis C 2005 Clinical Guidelines Summary of The: New York State Department of Health Clinical Guidelines for the Medical Management of Hepatitis C
Hepatitis C 2005 Clinical Guidelines Summary of the: New York State Department of Health Clinical Guidelines for the Medical Management of Hepatitis C Inside: Key Features of Viral Hepatitis A,B and C 1 Natural Course of HCV Infection 2 Persons at Risk for HCV Infection 3 Sources of HCV Infection 4 Counseling Prior To Testing 5 Screening for HCV Algorithm 6 Laboratory Testing for HCV 7 Interpretation of HCV Test Results 8 Post Exposure Screening for HCV 9 Counseling After Testing 10 Treating HCV Patients 11 Medical Management of HCV Positive Patients 12 References and Internet Resources 13 Hepatitis C Virus The Hidden Epidemic The Burden of HCV • 3 million Americans are chronically infected with the • 8-10,000 deaths a year are caused by HCV. Hepatitis C virus (HCV). • HCV is the leading cause for liver transplants and • 342,000 New Yorkers are estimated to be infected chronic liver disease. with HCV. • HCV deaths will increase four-fold to 38,000, by • A majority of the people infected with HCV do not the year 2010. know they have it. • Years of life lost to Hepatitis C (2001-2019) • Thousands of people go undetected each year—due 3.1 million years to inadequate risk assessment, under-screening and confusion about the use of diagnostic tests. • Cost of premature disability and death (2010-2109) $75.5 billion Hepatitis C Virus • Direct medical costs in absentee losses due to Hepatitis C $750 million/ year • HCV is a blood-borne disease transmitted by • Total medical expenditures for persons with HCV blood-to-blood contact. -

Host Cell Functions in Vesicular Stomatitis Virus Replication Phat X
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Veterinary and Biomedical Science Veterinary and Biomedical Sciences, Department of 2016 Host Cell Functions In Vesicular Stomatitis Virus Replication Phat X. Dinh University of Nebraska - Lincoln Anshuman Das University of Nebraska-Lincoln Asit K. Pattnaik University of Nebraska - Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/vetscipapers Part of the Biochemistry, Biophysics, and Structural Biology Commons, Cell and Developmental Biology Commons, Immunology and Infectious Disease Commons, Medical Sciences Commons, Veterinary Microbiology and Immunobiology Commons, and the Veterinary Pathology and Pathobiology Commons Dinh, Phat X.; Das, Anshuman; and Pattnaik, Asit K., "Host Cell Functions In Vesicular Stomatitis Virus Replication" (2016). Papers in Veterinary and Biomedical Science. 237. https://digitalcommons.unl.edu/vetscipapers/237 This Article is brought to you for free and open access by the Veterinary and Biomedical Sciences, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Papers in Veterinary and Biomedical Science by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published in Biology and Pathogenesis of Rhabdo and Filoviruses Copyright © 2016 World Scientific Publishing Co Pte Ltd digitalcommons.unl.edu HOST CELL FUNCTIONS IN VESICULAR STOMATITIS VIRUS REPLICATION Phat X. Dinh, Anshuman Das, and Asit K. Pattnaik School of Veterinary Medicine and Biomedical Sciences and the Nebraska Center for Virology, University of Nebraska-Lincoln, 109 Morrison Life Science Research Center, 4240 Fair Street, Lincoln, * Nebraska 68583, USA E-mail: [email protected] Summary Vesicular stomatitis virus (VSV), the prototypic rhabdovirus, has been used as an excellent paradigm for understanding the mechanisms of virus replication, pathogenesis, host response to virus infection and also for myriads of studies on cellular and molecular biology. -

Hepatitis C Virus Infection and Human Pancreatic ß-Cell Dysfunction
Pathophysiology/Complications BRIEF REPORT Hepatitis C Virus Infection and Human Pancreatic -Cell Dysfunction 1 1 MATILDE MASINI, MD SILVIA DEL GUERRA, PHD showing the characteristic insulin gran- 2 1 DANIELA CAMPANI, MD MARCO BUGLIANI, PHD ules and normally preserved mitochon- 3 1 UGO BOGGI, MD SCILLA TORRI, PHD  2 1 dria. In -cells from HCV-positive MICHELE MENICAGLI, MD STEFANO DEL PRATO, MD 1 3 pancreases, the presence of virus-like par- NICOLA FUNEL, MD FRANCO MOSCA, MD 1 4 ticles was observed, mainly close to the MARIA POLLERA, MD FRANCO FILIPPONI, MD 1 1 membranes of Golgi apparatus, which, in ROBERTO LUPI, PHD PIERO MARCHETTI, MD, PHD turn, appeared hyperplastic and dilated (Fig. 1D). The mitochondria appeared round-shaped with dispersed matrix and fragmented cristae (Fig. 1D). Additional Ϯ 2 any patients with chronic hepati- and 2 women, BMI 25.8 1.6 kg/m ) -cell changes were observed at the Ϯ tis C virus (HCV) develop type 2 and 10 HCV-negative (age 67 9 years, level of rough endoplasmic reticulum, Ϯ diabetes (1). This prevalence is 6 men and 4 women, BMI 26.8 2.0 which showed long and dilated tubular M 2 much higher than that observed in the kg/m ) donors were harvested and stud- membranes, with numerous electron- general population and in patients with ied with the approval of our local ethics dense ribosomes bound to the latter (not other chronic liver diseases such as hepa- committee. Histological studies were shown). These morphological changes titis B virus, alcoholic liver disease, and performed by immunohistochemistry were accompanied by reduced in vitro primary biliary cirrhosis. -

Hepatitis a Transmitted by Food
INVITED ARTICLE FOOD SAFETY David Acheson, Section Editor Hepatitis A Transmitted by Food Anthony E. Fiore Division of Viral Hepatitis, Centers for Disease Control and Prevention, Atlanta Hepatitis A is caused by hepatitis A virus (HAV). Transmission occurs by the fecal-oral route, either by direct contact with an HAV-infected person or by ingestion of HAV-contaminated food or water. Foodborne or waterborne hepatitis A outbreaks are relatively uncommon in the United States. However, food handlers with hepatitis A are frequently identified, and evaluation of the need for immunoprophylaxis and implementation of control measures are a considerable burden on public health resources. In addition, HAV-contaminated food may be the source of hepatitis A for an unknown proportion of persons whose source of infection is not identified. FEATURES OF HEPATITIS A and 40%–70% are jaundiced [6]. Children and occasionally young adults can also have inapparent infection, in which Hepatitis A virus (HAV) is classified as a picornavirus. Primates symptoms and elevation of ALT levels are absent but serocon are the only natural host [1]. There is only 1 HAV serotype, version occurs [7]. and immunity after infection is lifelong [2]. After ingestion, Hepatitis A begins with symptoms such as fever, anorexia, uptake in the gastrointestinal tract, and subsequent replication nausea, vomiting, diarrhea, myalgia, and malaise. Jaundice, in the liver, HAV is excreted in bile, and high concentrations dark-colored urine, or light-colored stools might be present at are found in stool specimens. Transmission occurs by the fecal- onset or might follow constitutional symptoms within a few oral route, either by direct contact with an HAV-infected person days.