GC-MS Metabolomics Reveals Metabolic Differences of the Farmed
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Kyoto Encyclopedia of Genes and Genomes (KEGG)
Kyoto Encyclopedia of Genes and Genome Minoru Kanehisa Institute for Chemical Research, Kyoto University HFSPO Workshop, Strasbourg, November 18, 2016 The KEGG Databases Category Database Content PATHWAY KEGG pathway maps Systems information BRITE BRITE functional hierarchies MODULE KEGG modules KO (KEGG ORTHOLOGY) KO groups for functional orthologs Genomic information GENOME KEGG organisms, viruses and addendum GENES / SSDB Genes and proteins / sequence similarity COMPOUND Chemical compounds GLYCAN Glycans Chemical information REACTION / RCLASS Reactions / reaction classes ENZYME Enzyme nomenclature DISEASE Human diseases DRUG / DGROUP Drugs / drug groups Health information ENVIRON Health-related substances (KEGG MEDICUS) JAPIC Japanese drug labels DailyMed FDA drug labels 12 manually curated original DBs 3 DBs taken from outside sources and given original annotations (GENOME, GENES, ENZYME) 1 computationally generated DB (SSDB) 2 outside DBs (JAPIC, DailyMed) KEGG is widely used for functional interpretation and practical application of genome sequences and other high-throughput data KO PATHWAY GENOME BRITE DISEASE GENES MODULE DRUG Genome Molecular High-level Practical Metagenome functions functions applications Transcriptome etc. Metabolome Glycome etc. COMPOUND GLYCAN REACTION Funding Annual budget Period Funding source (USD) 1995-2010 Supported by 10+ grants from Ministry of Education, >2 M Japan Society for Promotion of Science (JSPS) and Japan Science and Technology Agency (JST) 2011-2013 Supported by National Bioscience Database Center 0.8 M (NBDC) of JST 2014-2016 Supported by NBDC 0.5 M 2017- ? 1995 KEGG website made freely available 1997 KEGG FTP site made freely available 2011 Plea to support KEGG KEGG FTP academic subscription introduced 1998 First commercial licensing Contingency Plan 1999 Pathway Solutions Inc. -

Precise Generation of Systems Biology Models from KEGG Pathways Clemens Wrzodek*,Finjabuchel,¨ Manuel Ruff, Andreas Drager¨ and Andreas Zell
Wrzodek et al. BMC Systems Biology 2013, 7:15 http://www.biomedcentral.com/1752-0509/7/15 METHODOLOGY ARTICLE OpenAccess Precise generation of systems biology models from KEGG pathways Clemens Wrzodek*,FinjaBuchel,¨ Manuel Ruff, Andreas Drager¨ and Andreas Zell Abstract Background: The KEGG PATHWAY database provides a plethora of pathways for a diversity of organisms. All pathway components are directly linked to other KEGG databases, such as KEGG COMPOUND or KEGG REACTION. Therefore, the pathways can be extended with an enormous amount of information and provide a foundation for initial structural modeling approaches. As a drawback, KGML-formatted KEGG pathways are primarily designed for visualization purposes and often omit important details for the sake of a clear arrangement of its entries. Thus, a direct conversion into systems biology models would produce incomplete and erroneous models. Results: Here, we present a precise method for processing and converting KEGG pathways into initial metabolic and signaling models encoded in the standardized community pathway formats SBML (Levels 2 and 3) and BioPAX (Levels 2 and 3). This method involves correcting invalid or incomplete KGML content, creating complete and valid stoichiometric reactions, translating relations to signaling models and augmenting the pathway content with various information, such as cross-references to Entrez Gene, OMIM, UniProt ChEBI, and many more. Finally, we compare several existing conversion tools for KEGG pathways and show that the conversion from KEGG to BioPAX does not involve a loss of information, whilst lossless translations to SBML can only be performed using SBML Level 3, including its recently proposed qualitative models and groups extension packages. -

Table of Codes for Each Court of Each Level
Table of Codes for Each Court of Each Level Corresponding Type Chinese Court Region Court Name Administrative Name Code Code Area Supreme People’s Court 最高人民法院 最高法 Higher People's Court of 北京市高级人民 Beijing 京 110000 1 Beijing Municipality 法院 Municipality No. 1 Intermediate People's 北京市第一中级 京 01 2 Court of Beijing Municipality 人民法院 Shijingshan Shijingshan District People’s 北京市石景山区 京 0107 110107 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Haidian District of Haidian District People’s 北京市海淀区人 京 0108 110108 Beijing 1 Court of Beijing Municipality 民法院 Municipality Mentougou Mentougou District People’s 北京市门头沟区 京 0109 110109 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Changping Changping District People’s 北京市昌平区人 京 0114 110114 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Yanqing County People’s 延庆县人民法院 京 0229 110229 Yanqing County 1 Court No. 2 Intermediate People's 北京市第二中级 京 02 2 Court of Beijing Municipality 人民法院 Dongcheng Dongcheng District People’s 北京市东城区人 京 0101 110101 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Xicheng District Xicheng District People’s 北京市西城区人 京 0102 110102 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Fengtai District of Fengtai District People’s 北京市丰台区人 京 0106 110106 Beijing 1 Court of Beijing Municipality 民法院 Municipality 1 Fangshan District Fangshan District People’s 北京市房山区人 京 0111 110111 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Daxing District of Daxing District People’s 北京市大兴区人 京 0115 -

List of Main Production Facilities of ALDI North's Suppliers for Apparel
List of Main Production Facilities of ALDI North‘s Suppliers for Apparel, Home Textiles and Shoes Version March 2020 Produktionsstättenliste | März 2018 | Seite 0/17 Name Address Number of Employees Commodity Group Bangladesh AKH Eco Apparels Ltd. 495 Balitha, Shahbelishwer, Dhamrai > 5000 Apparel Chantik Garments Ltd. Kumkumari, Gouripur, Ashulia, Savar 2001 - 3000 Apparel Crossline Factory Ltd. 25 Vadam, Uttarpara, Nishatnagar, Tongi 1001 - 2000 Apparel Plot No. 45, 48, 49, 51 & 52; Holding No. 3/C Vadam, Post office: Nishatnagar, Crossline Knit Fabrics Ltd. 1001 - 2000 Apparel Tongi Designtex Knitwear Ltd. Beraiderchala, Sreepur, Gazipur 1001 - 2000 Apparel Disari Apparels (Pvt) Ltd. B-193 BSCIC Industrial Estate, Tongi 0 - 500 Apparel EH Fabrics Ltd. 221-225 Sataish Road, Khortoil, Tongi 1001 - 2000 Apparel Elaine Apparels Ltd. Charabagh, Amtala, Ashulia, Savar-1340 1001 - 2000 Apparel Eurotex Knitwear Ltd. Nayamati, Kutubpur, Fatullah 1001 - 2000 Apparel Faiza Industries Ltd. 425 Baimail, Konabari, P.O.-Kashem Cotton Mill, Thana-Joydebpur 1001 - 2000 Apparel Florence Fabrics Ltd. Bymile, Konabari, Gazipur 1346, Dhaka 501 - 1000 Apparel Fortis Garments Ltd. 100/1 Block-B, East Chandra, Shaheed Mosharraf Hossain Road, Kaliakoir 1001 - 2000 Apparel Habitus Fashion Ltd. Gajaria Para, Bahwal Mirzapur, Gazipur Sadar 2001 - 3000 Apparel Ilmeeyat Apparels Ltd. Jamirdia, Hobirbari, Valuka > 5000 Apparel Impress Fashion Ltd. Dag No. 2224 Matuail, Konapara, Katherpool, Jatrabari 2001 - 3000 Apparel J.L. Sweaters Ltd. Zirabo, P.O.-Ashulia, Thana-Ashulia, Savar 1001 - 2000 Apparel KA Design Ltd. Vill: Boro Beraid, P.S: Badda, Dhaka 1212 1001 - 2000 Apparel KC Bottom And Shirt Wear Company K. C. Industrial Park, Ratuti, Katchkura, Uttarkhan, Uttara, Dhaka 501 - 1000 Apparel BSCIC Hosiery Industrial Estate, Road No. -

2018 Annual Report CONTENTS
2018 Annual Report CONTENTS Important Notice 2 Section I Definitions 4 Section II Corporate Profile and Main Financial Indicators 6 Section III Corporate Business Summary 13 Section IV Report of the Board of Directors 17 Section V Major Events 45 Section VI Change of Ordinary Shares and Shareholders 77 Section VII Directors, Supervisors, Senior Management and Staff 85 Section VIII Corporate Governance Structure and Governance 98 Report Section IX Environmental, Social and Governance Report 126 Section X Report of the Supervisory Committee 166 Section XI Independent Auditor’s Report and Consolidated 168 Financial Statements Section XII Documents Available for Inspection 273 Appendix 281 ——Information Disclosure Index 274 ——Profile of the Highways 281 ——Vehicles Classification and Toll Rates 284 ——Toll Rates Applicable to Goods Vehicles 285 with Reference to Their Weight ——The Additional Toll Rates Applicable to Lixuan Expressway 286 ——National Trunk Highways 287 ——The Map of the Highway Network of Anhui Province 288 Important Notice 1. The Board of the Directors, the Supervisory Committee and the Directors, Supervisors and the Senior Management of the Company hereby warrant that the contents of the annual report are true, accurate and complete, and that there are no false accounts, misleading statements or significant omissions of information and jointly and severally accept the legal responsibility. 2. Absence of Directors Role of the Name of the Name of absent director absent director Reason of absence alternate director Non-executive director Yang Xudong Business commitment Du Jian Independent non-executive director Jiang Jun Business commitment Kong Yat Fan 3. PricewaterhouseCoopers Zhong Tian LLP (PRC Accountant) and PricewaterhouseCoopers (Hong Kong Accountant) have issued standard unqualified audit opinions on the consolidated financial statements of the Company. -
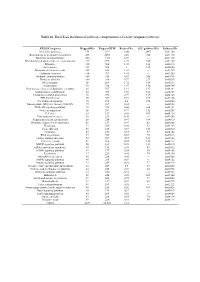
Table 1S. the KEGG Biochemical Pathways Categorization of LA Lily Unigenes Pathways
Table 1S. The KEGG biochemical pathways categorization of LA lily unigenes pathways. KEGG Categories Mapped-KO Unigene-NUM Ratio of No. ALL pathway KO Pathway-ID Metabolic pathways 954 4337 8.66 2067 ko01100 Biosynthesis of secondary metabolites 403 2285 4.56 720 ko01110 Biosynthesis of antibiotics 206 1147 2.29 --- ko01130 Microbial metabolism in diverse environments 157 995 1.99 720 ko01120 Ribosome 122 504 1.01 142 ko03010 Spliceosome 107 502 1 115 ko03040 Biosynthesis of amino acids 105 614 1.23 --- ko01230 Carbon metabolism 104 707 1.41 --- ko01200 Oxidative phosphorylation 100 335 0.67 206 ko00190 Purine metabolism 100 386 0.77 237 ko00230 RNA transport 98 861 1.72 134 ko03013 Endocytosis 92 736 1.47 138 ko04144 Protein processing in endoplasmic reticulum 88 567 1.13 137 ko04141 homologous recombination 84 933 1.86 144 ko05169 Ubiquitin mediated proteolysis 78 396 0.79 119 ko04120 HTLV-I infection 76 367 0.73 199 ko05166 Pyrimidine metabolism 76 298 0.6 150 ko00240 Non-alcoholic fatty liver disease (NAFLD) 72 209 0.42 --- ko04932 PI3K-Akt signaling pathway 71 328 0.66 226 ko04151 Viral carcinogenesis 68 387 0.77 132 ko05203 Cell cycle 63 339 0.68 103 ko04110 Proteoglycans in cancer 58 229 0.46 --- ko05205 Regulation of actin cytoskeleton 58 234 0.47 144 ko04810 Ribosome biogenesis in eukaryotes 56 237 0.47 82 ko03008 Phagosome 54 280 0.56 93 ko04145 Focal adhesion 53 185 0.37 133 ko04510 Lysosome 53 253 0.51 99 ko04142 RNA degradation 53 305 0.61 70 ko03018 Herpes simplex infection 52 257 0.51 121 ko05168 Cell cycle - yeast 51 260 -

A Tool for Retrieving Pathway Genes from KEGG Pathway Database
bioRxiv preprint doi: https://doi.org/10.1101/416131; this version posted September 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. KPGminer: A tool for retrieving pathway genes from KEGG pathway database A. K. M. Azad School of Biotechnology and Biomolecular Sciences, University of NSW, Chancellery Walk, Kensington, 2033, Australia Abstract Pathway analysis is a very important aspect in computational systems bi- ology as it serves as a crucial component in many computational pipelines. KEGG is one of the prominent databases that host pathway information associated with various organisms. In any pathway analysis pipelines, it is also important to collect and organize the pathway constituent genes for which a tool to automatically retrieve that would be a useful one to the practitioners. In this article, I present KPGminer, a tool that retrieves the constituent genes in KEGG pathways for various organisms and or- ganizes that information suitable for many downstream pathway analysis pipelines. We exploited several KEGG web services using REST APIs, par- ticularly GET and LIST methods to request for the information retrieval which is available for developers. Moreover, KPGminer can operate both for a particular pathway (single mode) or multiple pathways (batch mode). Next, we designed a crawler to extract necessary information from the re- sponse and generated outputs accordingly. KPGminer brings several key features including organism-specific and pathway-specific extraction of path- way genes from KEGG and always up-to-date information. -

Appendix Iii Property Valuation Report
THIS DOCUMENT IS IN DRAFT FORM, INCOMPLETE AND SUBJECT TO CHANGE AND THAT THE INFORMATION MUST BE READ IN CONJUNCTION WITH THE SECTION HEADED ‘‘WARNING’’ ON THE COVER OF THIS DOCUMENT. APPENDIX III PROPERTY VALUATION REPORT The following is the text of a letter, summary of values and valuation report prepared for the purpose of incorporation in this document received from Savills Valuation and Professional Services Limited, an independent valuer, in connection with their opinion of values of the property interests held by the Group as at 28 February 2021. The Directors Shiliu Investment Group Limited Building 1, Anar Center, No. 88 Liuxiang Road, Fengtai District, Beijing, The People’s Republic of China Savills Valuation and Professional Services Limited [Date] Room 1208, 1111 King’s Road, Taikoo Shing Hong Kong T: (852) 2801 6100 F: (852) 2530 0756 EA Licence: C-023750 savills.com Dear Sirs, INSTRUCTIONS In accordance with your instructions to us to value the properties (the ‘‘properties’’) situated in the People’s Republic of China (the ‘‘PRC’’) in which Shiliu Investment Group Limited (石榴投資集 團有限公司) (the ‘‘Company’’) and its subsidiaries (hereinaftertogetherreferredtoasthe‘‘Group’’) have interests, we confirm that we have carried out inspections, made relevant enquiries and obtained such further information as we consider necessary for the purpose of providing you with our opinion of market values of the properties as at 28 February 2021 (the ‘‘valuation date’’) for incorporation in a [REDACTED] Document. IDENTIFICATION AND STATUS OF THE VALUER The subject valuation exercise is handled by Mr. Anthony C.K. Lau, who is a Director of Savills Valuation and Professional Services Limited (‘‘SVPSL’’) and a corporate member of HKIS with over 28 years’ experience in valuation of properties in the PRC and has sufficient knowledge of the relevant market, the skills and understanding to handle the subject valuation exercise competently. -
2019 Public Supplier List
2019 Public Supplier List The list of suppliers includes original design manufacturers (ODMs), final assembly, and direct material suppliers. The volume and nature of business conducted with suppliers shifts as business needs change. This list represents a snapshot covering at least 95% of Dell’s spend during fiscal year 2020. Corporate Profile Address Dell Products / Supplier Type Sustainability Site GRI Procurement Category Compal 58 First Avenue, Alienware Notebook Final Assembly and/ Compal YES Comprehensive Free Trade Latitude Notebook or Original Design Zone, Kunshan, Jiangsu, Latitude Tablet Manufacturers People’s Republic Of China Mobile Precision Notebook XPS Notebook Poweredge Server Compal 8 Nandong Road, Pingzhen IoT Desktop Final Assembly and/ Compal YES District, Taoyuan, Taiwan IoT Embedded PC or Original Design Latitude Notebook Manufacturers Latitude Tablet Mobile Precision Notebook Compal 88, Sec. 1, Zongbao Inspiron Notebook Final Assembly and/ Compal YES Avenue, Chengdu Hi-tech Vostro Notebook or Original Design Comprehensive Bonded Zone, Manufacturers Shuangliu County, Chengdu, Sichuan, People’s Republic of China Public Supplier List 2019 | Published November 2020 | 01 of 40 Corporate Profile Address Dell Products / Supplier Type Sustainability Site GRI Procurement Category Dell 2366-2388 Jinshang Road, Alienware Desktop Final Assembly and/ Dell YES Information Guangdian, Inspiron Desktop or Original Design Xiamen, Fujian, People’s Optiplex All in One Manufacturers Republic of China Optiplex Desktop Precision Desktop -

Downregulated Developmental Processes in the Postnatal Right
www.nature.com/cddiscovery ARTICLE OPEN Downregulated developmental processes in the postnatal right ventricle under the influence of a volume overload ✉ ✉ ✉ Chunxia Zhou1,6, Sijuan Sun2,6, Mengyu Hu3, Yingying Xiao1, Xiafeng Yu1 , Lincai Ye 1,4,5 and Lisheng Qiu 1 © The Author(s) 2021 The molecular atlas of postnatal mouse ventricular development has been made available and cardiac regeneration is documented to be a downregulated process. The right ventricle (RV) differs from the left ventricle. How volume overload (VO), a common pathologic state in children with congenital heart disease, affects the downregulated processes of the RV is currently unclear. We created a fistula between the abdominal aorta and inferior vena cava on postnatal day 7 (P7) using a mouse model to induce a prepubertal RV VO. RNAseq analysis of RV (from postnatal day 14 to 21) demonstrated that angiogenesis was the most enriched gene ontology (GO) term in both the sham and VO groups. Regulation of the mitotic cell cycle was the second-most enriched GO term in the VO group but it was not in the list of enriched GO terms in the sham group. In addition, the number of Ki67-positive cardiomyocytes increased approximately 20-fold in the VO group compared to the sham group. The intensity of the vascular endothelial cells also changed dramatically over time in both groups. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the downregulated transcriptome revealed that the peroxisome proliferators-activated receptor (PPAR) signaling pathway was replaced by the cell cycle in the top-20 enriched KEGG terms because of the VO. -

Transcriptome Analysis of the Regulatory Mechanism of Foxo on Wing Dimorphism in the Brown Planthopper, Nilaparvata Lugens (Hemiptera: Delphacidae)
insects Article Transcriptome Analysis of the Regulatory Mechanism of FoxO on Wing Dimorphism in the Brown Planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) Nan Xu 1, Sheng-Fei Wei 1 and Hai-Jun Xu 1,2,3,* 1 State Key Laboratory of Rice Biology, Zhejiang University, Hangzhou 310058, China; [email protected] (N.X.); [email protected] (S.-F.W.) 2 Ministry of Agriculture Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Zhejiang University, Hangzhou 310058, China 3 Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China * Correspondence: [email protected]; Tel.: +86-571-88982996 Simple Summary: The brown planthopper (BPH) Nilaparvata lugens can develop into either long- winged or short-winged adults depending on environmental stimuli received during larval stages. The transcription factor NlFoxO serves as a key regulator determining alternative wing morphs in BPH, but the underlying molecular mechanism is largely unknown. Here, we investigated the transcriptomic profile of forewing and hindwing buds across the 5th-instar stage, the wing-morph decision stage. Our results indicated that NlFoxO modulated the developmental plasticity of wing buds mainly by regulating the expression of cell proliferation-associated genes. Abstract: The brown planthopper (BPH), Nilaparvata lugens, can develop into either short-winged (SW) or long-winged (LW) adults according to environmental conditions, and has long served as a model organism for exploring the mechanisms of wing polyphenism in insects. The transcription Citation: Xu, N.; Wei, S.-F.; Xu, H.-J. factor NlFoxO acts as a master regulator that directs the development of either SW or LW morphs, Transcriptome Analysis of the but the underlying molecular mechanism is largely unknown. -

"Using the KEGG Database Resource". In: Current Protocols in Bioinformatics
Using the KEGG Database Resource UNIT 1.12 KEGG (Kyoto Encyclopedia of Genes and Genomes) is a bioinformatics resource for understanding biological function from a genomic perspective. It is a multispecies, integrated resource consisting of genomic, chemical, and network information with cross-references to numerous outside databases and containing a complete set of building blocks (genes and molecules) and wiring diagrams (biological pathways) to represent cellular functions (Kanehisa et al., 2004). In this unit, protocols are described for using the five major KEGG resources: PATHWAY, GENES, SSDB, EXPRESSION, and LIGAND. The KEGG PATHWAY database (see Basic Protocols 1 to 4) consists of a user-friendly tool for analyzing the network of protein and small-molecule interactions that occur in the cells of various organisms. KEGG GENES (see Basic Protocols 5 and 6) provides access to the collection of gene data organized so as to be accessible via text searches, from the PATHWAY database, or via cross-species orthology searches. The KEGG Sequence Similiarity Database (SSDB; see Basic Protocols 7 to 9) consists of a precomputed database of all-versus-all Smith- Waterman similarity scores among all genes in KEGG GENES, enabling relationships between homologs to be easily visualized on the pathway and genome maps or viewed as clusters of orthologous genes. The KEGG EXPRESSION database (see Basic Protocols 10 to 14) contains both data and tools for analyzing gene expression data. User-defined data such as microarray experiments may also be uploaded for analysis using the tools available in KEGG EXPRESSION. Finally, KEGG LIGAND (see Basic Protocols 16 to 19) is a database of small molecules, their structures, and information relating to the enzymes that act on them.