Mechanics and Structural Stability of the Collagen Triple Helix
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Flexible Fluidic Actuators for Soft Robotic Applications
University of Wollongong Research Online University of Wollongong Thesis Collection 2017+ University of Wollongong Thesis Collections 2019 Flexible Fluidic Actuators for Soft Robotic Applications Weiping Hu Follow this and additional works at: https://ro.uow.edu.au/theses1 University of Wollongong Copyright Warning You may print or download ONE copy of this document for the purpose of your own research or study. The University does not authorise you to copy, communicate or otherwise make available electronically to any other person any copyright material contained on this site. You are reminded of the following: This work is copyright. Apart from any use permitted under the Copyright Act 1968, no part of this work may be reproduced by any process, nor may any other exclusive right be exercised, without the permission of the author. Copyright owners are entitled to take legal action against persons who infringe their copyright. A reproduction of material that is protected by copyright may be a copyright infringement. A court may impose penalties and award damages in relation to offences and infringements relating to copyright material. Higher penalties may apply, and higher damages may be awarded, for offences and infringements involving the conversion of material into digital or electronic form. Unless otherwise indicated, the views expressed in this thesis are those of the author and do not necessarily represent the views of the University of Wollongong. Research Online is the open access institutional repository for the University of -

Environmentally Controlled Curvature of Single Collagen Proteins
Environmentally controlled curvature of single collagen proteins Naghmeh Rezaei, Aaron Lyons and Nancy R. Forde* Department of Physics Simon Fraser University 8888 University Drive Burnaby, BC V5A 1S6 CANADA *Corresponding author. 778-782-3161. [email protected]. ORCID: 0000-0002-5479-7073 - 1 - ABSTRACT The predominant structural protein in vertebrates is collagen, which plays a key role in extracellular matrix and connective tissue mechanics. Despite its prevalence and physical importance in biology, the mechanical properties of molecular collagen are far from established. The flexibility of its triple helix is unresolved, with descriptions from different experimental techniques ranging from flexible to semirigid. Furthermore, it is unknown how collagen type (homo- vs. heterotrimeric) and source (tissue-derived vs. recombinant) influence flexibility. Using SmarTrace, a chain tracing algorithm we devised, we performed statistical analysis of collagen conformations collected with atomic force microscopy (AFM) to Our results show that types I, II and III collagens the key fibrillar varieties exhibit molecular flexibilities that are very similar. However, collagen conformations are strongly modulated by salt, transitioning from compact to extended as KCl concentration increases, in both neutral and acidic pH. While analysis with a standard worm-like chain model suggests that the persistence length of collagen can attain almost any value within the literature range, closer inspection reveals that this modulation to changes in flexibility, but rather arises from the induction of curvature (either intrinsic or induced by interactions with the mica surface). By modifying standard polymer theory to include innate curvature, we show that collagen behaves as an equilibrated curved worm-like chain (cWLC) in two dimensions. -
Repair, Regeneration, and Fibrosis Gregory C
91731_ch03 12/8/06 7:33 PM Page 71 3 Repair, Regeneration, and Fibrosis Gregory C. Sephel Stephen C. Woodward The Basic Processes of Healing Regeneration Migration of Cells Stem cells Extracellular Matrix Cell Proliferation Remodeling Conditions That Modify Repair Cell Proliferation Local Factors Repair Repair Patterns Repair and Regeneration Suboptimal Wound Repair Wound Healing bservations regarding the repair of wounds (i.e., wound architecture are unaltered. Thus, wounds that do not heal may re- healing) date to physicians in ancient Egypt and battle flect excess proteinase activity, decreased matrix accumulation, Osurgeons in classic Greece. The liver’s ability to regenerate or altered matrix assembly. Conversely, fibrosis and scarring forms the basis of the Greek myth involving Prometheus. The may result from reduced proteinase activity or increased matrix clotting of blood to prevent exsanguination was recognized as accumulation. Whereas the formation of new collagen during the first necessary event in wound healing. At the time of the repair is required for increased strength of the healing site, American Civil War, the development of “laudable pus” in chronic fibrosis is a major component of diseases that involve wounds was thought to be necessary, and its emergence was not chronic injury. appreciated as a symptom of infection but considered a positive sign in the healing process. Later studies of wound infection led The Basic Processes of Healing to the discovery that inflammatory cells are primary actors in the repair process. Although scurvy (see Chapter 8) was described in Many of the basic cellular and molecular mechanisms necessary the 16th century by the British navy, it was not until the 20th for wound healing are found in other processes involving dynamic century that vitamin C (ascorbic acid) was found to be necessary tissue changes, such as development and tumor growth. -

Collagen VI-Related Myopathy
Collagen VI-related myopathy Description Collagen VI-related myopathy is a group of disorders that affect skeletal muscles (which are the muscles used for movement) and connective tissue (which provides strength and flexibility to the skin, joints, and other structures throughout the body). Most affected individuals have muscle weakness and joint deformities called contractures that restrict movement of the affected joints and worsen over time. Researchers have described several forms of collagen VI-related myopathy, which range in severity: Bethlem myopathy is the mildest, an intermediate form is moderate in severity, and Ullrich congenital muscular dystrophy is the most severe. People with Bethlem myopathy usually have loose joints (joint laxity) and weak muscle tone (hypotonia) in infancy, but they develop contractures during childhood, typically in their fingers, wrists, elbows, and ankles. Muscle weakness can begin at any age but often appears in childhood to early adulthood. The muscle weakness is slowly progressive, with about two-thirds of affected individuals over age 50 needing walking assistance. Older individuals may develop weakness in respiratory muscles, which can cause breathing problems. Some people with this mild form of collagen VI-related myopathy have skin abnormalities, including small bumps called follicular hyperkeratosis on the arms and legs; soft, velvety skin on the palms of the hands and soles of the feet; and abnormal wound healing that creates shallow scars. The intermediate form of collagen VI-related myopathy is characterized by muscle weakness that begins in infancy. Affected children are able to walk, although walking becomes increasingly difficult starting in early adulthood. They develop contractures in the ankles, elbows, knees, and spine in childhood. -

The Beneficial Regulation of Extracellular Matrix
cosmetics Article The Beneficial Regulation of Extracellular Matrix and Heat Shock Proteins, and the Inhibition of Cellular Oxidative Stress Effects and Inflammatory Cytokines by 1α, 25 dihydroxyvitaminD3 in Non-Irradiated and Ultraviolet Radiated Dermal Fibroblasts Neena Philips *, Xinxing Ding, Pranathi Kandalai, Ilonka Marte, Hunter Krawczyk and Richard Richardson School of Natural Sciences, Fairleigh Dickinson University, Teaneck, NJ 07601, USA * Correspondence: [email protected] or [email protected] Received: 30 June 2019; Accepted: 20 July 2019; Published: 1 August 2019 Abstract: Intrinsic skin aging and photoaging, from exposure to ultraviolet (UV) radiation, are associated with altered regulation of genes associated with the extracellular matrix (ECM) and inflammation, as well as cellular damage from oxidative stress. The regulatory properties of 1α, 25dihydroxyvitamin D3 (vitamin D) include endocrine, ECM regulation, cell differentiation, photoprotection, and anti-inflammation. The goal of this research was to identify the beneficial effects of vitamin D in preventing intrinsic skin aging and photoaging, through its direct effects as well as its effects on the ECM, associated heat shock proteins (HSP-47, and -70), cellular oxidative stress effects, and inflammatory cytokines [interleukin (IL)-1 and IL-8] in non-irradiated, UVA-radiated, UVB-radiated dermal fibroblasts. With regard to the ECM, vitamin D stimulated type I collagen and inhibited cellular elastase activity in non-irradiated fibroblasts; and stimulated type I collagen and HSP-47, and inhibited elastin expression and elastase activity in UVA-radiated dermal fibroblasts. With regard to cellular protection, vitamin D inhibited oxidative damage to DNA, RNA, and lipids in non-irradiated, UVA-radiated and UVB-radiated fibroblasts, and, in addition, increased cell viability of UVB-radiated cells. -

Dietary Protein Restriction Rapidly Reduces Transforming Growth Factor
Proc. Natl. Acad. Sci. USA Vol. 88, pp. 9765-9769, November 1991 Medical Sciences Dietary protein restriction rapidly reduces transforming growth factor p1 expression in experimental glomerulonephritis (extraceliular matrix/transforming growth factor 8/glomerulonephritis) SEIYA OKUDA*, TAKAMICHI NAKAMURA*, TATSUO YAMAMOTO*, ERKKI RUOSLAHTIt, AND WAYNE A. BORDER*t *Division of Nephrology, University of Utah School of Medicine, Salt Lake City, UT 84132; and tCancer Research Center, La Jolla Cancer Research Foundation, La Jolla, CA 92037 Communicated by Eugene Roberts, August 19, 1991 (receivedfor review April 29, 1991) ABSTRACT Dietary protein restriction has been shown to TGF-/31 on both cell types is to regulate the synthesis of two slow the rate of loss of kidney function in humans with chondroitin/dermatan sulfate proteoglycans, biglycan and progressive glomerulosclerosis due to glomerulonephritis or decorin, both of which can bind TGF-P1 (23). In an experi- diabetes mellitus. A central feature of glomerulosclerosis is the mental model ofglomerulonephritis in the rat, we have found pathological accumulation of extracellular matrix within the a close association between elevated expression of the diseased glomeruli. Transforming growth factor j1 (TGF-.81) TGF-131 gene and the development of glomerulonephritis is known to have widespread regulatory effects on extracellular (10). Seven days after glomerular injury, at the time of matrix and has been implicated as a major cause of increased significant extracellular matrix accumulation, the glomeruli extracellular matrix synthesis and buildup of pathological showed a 5-fold increase in TGF-f31 mRNA and a nearly matrix within glomeruli in experimental glomerulonephritis. 50-fold increase in production ofbiglycan and decorin. -

Peptoid Residues Make Diverse, Hyperstable Collagen Triple Helices
Peptoid Residues Make Diverse, Hyperstable Collagen Triple Helices Julian L. Kessler1, Grace Kang1, Zhao Qin2, Helen Kang1, Frank G. Whitby3, Thomas E. Cheatham III4, Christopher P. Hill3, Yang Li1,*, and S. Michael Yu1,5 1Department of Biomedical Engineering, University of Utah, Salt Lake City, Utah 84112, USA 2Department of Civil & Environmental Engineering, Collagen of Engineering & Computer Science, Syracuse University, Syracuse, New York 13244, USA 3Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, UT 84112, USA 4Department of Medicinal Chemistry, College of Pharmacy, L. S. Skaggs Pharmacy Research Institute, University of Utah, Salt Lake City, Utah 84112, USA 5Department of Pharmaceutics and Pharmaceutical Chemistry, University of Utah, Salt Lake City, Utah 84112, USA *Corresponding Author: Yang Li ([email protected]) Abstract The triple-helical structure of collagen, responsible for collagen’s remarkable biological and mechanical properties, has inspired both basic and applied research in synthetic peptide mimetics for decades. Since non-proline amino acids weaken the triple helix, the cyclic structure of proline has been considered necessary, and functional collagen mimetic peptides (CMPs) with diverse sidechains have been difficult to produce. Here we show that N-substituted glycines (N-glys), also known as peptoid residues, exhibit a general triple-helical propensity similar to or greater than proline, allowing synthesis of thermally stable triple-helical CMPs with unprecedented sidechain diversity. We found that the N-glys stabilize the triple helix by sterically promoting the preorganization of individual CMP chains into the polyproline-II helix conformation. Our findings were supported by the crystal structures of two atomic-resolution N-gly-containing CMPs, as well as experimental and computational studies spanning more than 30 N-gly-containing peptides. -

Extracellular Matrix Grafts: from Preparation to Application (Review)
INTERNATIONAL JOURNAL OF MOleCular meDICine 47: 463-474, 2021 Extracellular matrix grafts: From preparation to application (Review) YONGSHENG JIANG1*, RUI LI1,2*, CHUNCHAN HAN1 and LIJIANG HUANG1 1Science and Education Management Center, The Affiliated Xiangshan Hospital of Wenzhou Medical University, Ningbo, Zhejiang 315700; 2School of Chemistry, Sun Yat-sen University, Guangzhou, Guangdong 510275, P.R. China Received July 30, 2020; Accepted December 3, 2020 DOI: 10.3892/ijmm.2020.4818 Abstract. Recently, the increasing emergency of traffic acci- Contents dents and the unsatisfactory outcome of surgical intervention are driving research to seek a novel technology to repair trau- 1. Introduction matic soft tissue injury. From this perspective, decellularized 2. ECM-G characterization matrix grafts (ECM-G) including natural ECM materials, and 3. Methods of decellularization treatments their prepared hydrogels and bioscaffolds, have emerged as 4. Removal of residual cellular components and chemicals possible alternatives for tissue engineering and regenerative 5. Application of ECM-P in regenerative medicine medicine. Over the past decades, several physical and chemical 6. Challenges and future outlook on ECM-P decellularization methods have been used extensively to deal 7. Conclusions with different tissues/organs in an attempt to carefully remove cellular antigens while maintaining the non-immunogenic ECM components. It is anticipated that when the decellular- 1. Introduction ized biomaterials are seeded with cells in vitro or incorporated into irregularly shaped defects in vivo, they can provide the The extracellular matrix (ECM) derived from organs/tissues is appropriate biomechanical and biochemical conditions for a complex, highly organized assembly of macromolecules with directing cell behavior and tissue remodeling. -
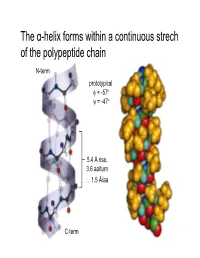
The Α-Helix Forms Within a Continuous Strech of the Polypeptide Chain
The α-helix forms within a continuous strech of the polypeptide chain N-term prototypical φ = -57 ° ψ = -47 ° 5.4 Å rise, 3.6 aa/turn ∴ 1.5 Å/aa C-term α-Helices have a dipole moment, due to unbonded and aligned N-H and C=O groups β-Sheets contain extended (β-strand) segments from separate regions of a protein prototypical φ = -139 °, ψ = +135 ° prototypical φ = -119 °, ψ = +113 ° (6.5Å repeat length in parallel sheet) Antiparallel β-sheets may be formed by closer regions of sequence than parallel Beta turn Figure 6-13 The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups Helix wheel The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups • Occurrence of proline and glycine • Interactions between ends of helix and aa R-groups His Glu N-term C-term -

Effects of Collagen-Derived Bioactive Peptides and Natural Antioxidant
www.nature.com/scientificreports OPEN Efects of collagen-derived bioactive peptides and natural antioxidant compounds on Received: 29 December 2017 Accepted: 19 June 2018 proliferation and matrix protein Published: xx xx xxxx synthesis by cultured normal human dermal fbroblasts Suzanne Edgar1, Blake Hopley1, Licia Genovese2, Sara Sibilla2, David Laight1 & Janis Shute1 Nutraceuticals containing collagen peptides, vitamins, minerals and antioxidants are innovative functional food supplements that have been clinically shown to have positive efects on skin hydration and elasticity in vivo. In this study, we investigated the interactions between collagen peptides (0.3–8 kDa) and other constituents present in liquid collagen-based nutraceuticals on normal primary dermal fbroblast function in a novel, physiologically relevant, cell culture model crowded with macromolecular dextran sulphate. Collagen peptides signifcantly increased fbroblast elastin synthesis, while signifcantly inhibiting release of MMP-1 and MMP-3 and elastin degradation. The positive efects of the collagen peptides on these responses and on fbroblast proliferation were enhanced in the presence of the antioxidant constituents of the products. These data provide a scientifc, cell-based, rationale for the positive efects of these collagen-based nutraceutical supplements on skin properties, suggesting that enhanced formation of stable dermal fbroblast-derived extracellular matrices may follow their oral consumption. Te biophysical properties of the skin are determined by the interactions between cells, cytokines and growth fac- tors within a network of extracellular matrix (ECM) proteins1. Te fbril-forming collagen type I is the predomi- nant collagen in the skin where it accounts for 90% of the total and plays a major role in structural organisation, integrity and strength2. -

Characterization of Collagen Fibers (I, III, IV) and Elastin of Normal and Neoplastic Canine Prostatic Tissues
veterinary sciences Article Characterization of Collagen Fibers (I, III, IV) and Elastin of Normal and Neoplastic Canine Prostatic Tissues Luis Gabriel Rivera Calderón 1, Priscila Emiko Kobayashi 2, Rosemeri Oliveira Vasconcelos 1, Carlos Eduardo Fonseca-Alves 3 and Renée Laufer-Amorim 2,* 1 Department of Veterinary Pathology, School of Agricultural and Veterinarian Sciences, São Paulo State University (Unesp), Jaboticabal, São Paulo 14884-900, Brazil; [email protected] (L.G.R.C.); [email protected] (R.O.V.) 2 Department of Veterinary Clinic, School of Veterinary Medicine and Animal Science, São Paulo State University (Unesp), Botucatu, São Paulo 18618-681, Brazil; [email protected] 3 Department of Veterinary Surgery and Anesthesiology, School of Veterinary Medicine and Animal Science, São Paulo State University (Unesp), Botucatu, São Paulo 18618-681, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-14-3880-2076 Received: 7 January 2019; Accepted: 25 February 2019; Published: 2 March 2019 Abstract: This study aimed to investigate collagen (Coll-I, III, IV) and elastin in canine normal prostate and prostate cancer (PC) using Picrosirius red (PSR) and Immunohistochemical (IHC) analysis. Eight normal prostates and 10 PC from formalin-fixed, paraffin-embedded samples were used. Collagen fibers area was analyzed with ImageJ software. The distribution of Coll-I and Coll-III was approximately 80% around prostatic ducts and acini, 15% among smooth muscle, and 5% surrounding blood vessels, in both normal prostate and PC. There was a higher median area of Coll-III in PC when compared to normal prostatic tissue (p = 0.001 for PSR and p = 0.05 for IHC). -

Y. Ma, X. Feng, J.A. Rogers, Y. Huang and Yihui
Lab on a Chip View Article Online CRITICAL REVIEW View Journal | View Issue Design and application of ‘J-shaped’ stress–strain Cite this: Lab Chip,2017,17,1689 behavior in stretchable electronics: a review Yinji Ma,ab Xue Feng,a John A. Rogers,c Yonggang Huangb and Yihui Zhang *a A variety of natural biological tissues (e.g., skin, ligaments, spider silk, blood vessel) exhibit ‘J-shaped’ stress–strain behavior, thereby combining soft, compliant mechanics and large levels of stretchability, with anatural‘strain-limiting’ mechanism to prevent damage from excessive strain. Synthetic materials with similar stress–strain behaviors have potential utility in many promising applications, such as tissue engineer- ing (to reproduce the nonlinear mechanical properties of real biological tissues) and biomedical devices (to enable natural, comfortable integration of stretchable electronics with biological tissues/organs). Recent advances in this field encompass developments of novel material/structure concepts, fabrication ap- proaches, and unique device applications. This review highlights five representative strategies, including de- signs that involve open network, wavy and wrinkled morphologies, helical layouts, kirigami and origami constructs, and textile formats. Discussions focus on the underlying ideas, the fabrication/assembly routes, Received 18th March 2017, and the microstructure–property relationships that are essential for optimization of the desired ‘J-shaped’ Accepted 21st April 2017 stress–strain responses. Demonstration applications provide examples of the use of these designs in de- formable electronics and biomedical devices that offer soft, compliant mechanics but with inherent robust- DOI: 10.1039/c7lc00289k ness against damage from excessive deformation. We conclude with some perspectives on challenges and rsc.li/loc opportunities for future research.