Peptoid Residues Make Diverse, Hyperstable Collagen Triple Helices
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Flexible Fluidic Actuators for Soft Robotic Applications
University of Wollongong Research Online University of Wollongong Thesis Collection 2017+ University of Wollongong Thesis Collections 2019 Flexible Fluidic Actuators for Soft Robotic Applications Weiping Hu Follow this and additional works at: https://ro.uow.edu.au/theses1 University of Wollongong Copyright Warning You may print or download ONE copy of this document for the purpose of your own research or study. The University does not authorise you to copy, communicate or otherwise make available electronically to any other person any copyright material contained on this site. You are reminded of the following: This work is copyright. Apart from any use permitted under the Copyright Act 1968, no part of this work may be reproduced by any process, nor may any other exclusive right be exercised, without the permission of the author. Copyright owners are entitled to take legal action against persons who infringe their copyright. A reproduction of material that is protected by copyright may be a copyright infringement. A court may impose penalties and award damages in relation to offences and infringements relating to copyright material. Higher penalties may apply, and higher damages may be awarded, for offences and infringements involving the conversion of material into digital or electronic form. Unless otherwise indicated, the views expressed in this thesis are those of the author and do not necessarily represent the views of the University of Wollongong. Research Online is the open access institutional repository for the University of -

Environmentally Controlled Curvature of Single Collagen Proteins
Environmentally controlled curvature of single collagen proteins Naghmeh Rezaei, Aaron Lyons and Nancy R. Forde* Department of Physics Simon Fraser University 8888 University Drive Burnaby, BC V5A 1S6 CANADA *Corresponding author. 778-782-3161. [email protected]. ORCID: 0000-0002-5479-7073 - 1 - ABSTRACT The predominant structural protein in vertebrates is collagen, which plays a key role in extracellular matrix and connective tissue mechanics. Despite its prevalence and physical importance in biology, the mechanical properties of molecular collagen are far from established. The flexibility of its triple helix is unresolved, with descriptions from different experimental techniques ranging from flexible to semirigid. Furthermore, it is unknown how collagen type (homo- vs. heterotrimeric) and source (tissue-derived vs. recombinant) influence flexibility. Using SmarTrace, a chain tracing algorithm we devised, we performed statistical analysis of collagen conformations collected with atomic force microscopy (AFM) to Our results show that types I, II and III collagens the key fibrillar varieties exhibit molecular flexibilities that are very similar. However, collagen conformations are strongly modulated by salt, transitioning from compact to extended as KCl concentration increases, in both neutral and acidic pH. While analysis with a standard worm-like chain model suggests that the persistence length of collagen can attain almost any value within the literature range, closer inspection reveals that this modulation to changes in flexibility, but rather arises from the induction of curvature (either intrinsic or induced by interactions with the mica surface). By modifying standard polymer theory to include innate curvature, we show that collagen behaves as an equilibrated curved worm-like chain (cWLC) in two dimensions. -

Structural Insights Into Membrane Fusion Mediated by Convergent Small Fusogens
cells Review Structural Insights into Membrane Fusion Mediated by Convergent Small Fusogens Yiming Yang * and Nandini Nagarajan Margam Department of Microbiology and Immunology, Dalhousie University, Halifax, NS B3H 4R2, Canada; [email protected] * Correspondence: [email protected] Abstract: From lifeless viral particles to complex multicellular organisms, membrane fusion is inarguably the important fundamental biological phenomena. Sitting at the heart of membrane fusion are protein mediators known as fusogens. Despite the extensive functional and structural characterization of these proteins in recent years, scientists are still grappling with the fundamental mechanisms underlying membrane fusion. From an evolutionary perspective, fusogens follow divergent evolutionary principles in that they are functionally independent and do not share any sequence identity; however, they possess structural similarity, raising the possibility that membrane fusion is mediated by essential motifs ubiquitous to all. In this review, we particularly emphasize structural characteristics of small-molecular-weight fusogens in the hope of uncovering the most fundamental aspects mediating membrane–membrane interactions. By identifying and elucidating fusion-dependent functional domains, this review paves the way for future research exploring novel fusogens in health and disease. Keywords: fusogen; SNARE; FAST; atlastin; spanin; myomaker; myomerger; membrane fusion 1. Introduction Citation: Yang, Y.; Margam, N.N. Structural Insights into Membrane Membrane fusion -
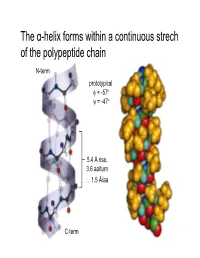
The Α-Helix Forms Within a Continuous Strech of the Polypeptide Chain
The α-helix forms within a continuous strech of the polypeptide chain N-term prototypical φ = -57 ° ψ = -47 ° 5.4 Å rise, 3.6 aa/turn ∴ 1.5 Å/aa C-term α-Helices have a dipole moment, due to unbonded and aligned N-H and C=O groups β-Sheets contain extended (β-strand) segments from separate regions of a protein prototypical φ = -139 °, ψ = +135 ° prototypical φ = -119 °, ψ = +113 ° (6.5Å repeat length in parallel sheet) Antiparallel β-sheets may be formed by closer regions of sequence than parallel Beta turn Figure 6-13 The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups Helix wheel The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups • Occurrence of proline and glycine • Interactions between ends of helix and aa R-groups His Glu N-term C-term -

Y. Ma, X. Feng, J.A. Rogers, Y. Huang and Yihui
Lab on a Chip View Article Online CRITICAL REVIEW View Journal | View Issue Design and application of ‘J-shaped’ stress–strain Cite this: Lab Chip,2017,17,1689 behavior in stretchable electronics: a review Yinji Ma,ab Xue Feng,a John A. Rogers,c Yonggang Huangb and Yihui Zhang *a A variety of natural biological tissues (e.g., skin, ligaments, spider silk, blood vessel) exhibit ‘J-shaped’ stress–strain behavior, thereby combining soft, compliant mechanics and large levels of stretchability, with anatural‘strain-limiting’ mechanism to prevent damage from excessive strain. Synthetic materials with similar stress–strain behaviors have potential utility in many promising applications, such as tissue engineer- ing (to reproduce the nonlinear mechanical properties of real biological tissues) and biomedical devices (to enable natural, comfortable integration of stretchable electronics with biological tissues/organs). Recent advances in this field encompass developments of novel material/structure concepts, fabrication ap- proaches, and unique device applications. This review highlights five representative strategies, including de- signs that involve open network, wavy and wrinkled morphologies, helical layouts, kirigami and origami constructs, and textile formats. Discussions focus on the underlying ideas, the fabrication/assembly routes, Received 18th March 2017, and the microstructure–property relationships that are essential for optimization of the desired ‘J-shaped’ Accepted 21st April 2017 stress–strain responses. Demonstration applications provide examples of the use of these designs in de- formable electronics and biomedical devices that offer soft, compliant mechanics but with inherent robust- DOI: 10.1039/c7lc00289k ness against damage from excessive deformation. We conclude with some perspectives on challenges and rsc.li/loc opportunities for future research. -

Protein Side-Chain Translocation Mutagenesis Via Incorporation of Peptoid Residues † Byoung-Chul Lee and Ronald N
ARTICLES pubs.acs.org/acschemicalbiology Protein Side-Chain Translocation Mutagenesis via Incorporation of Peptoid Residues † Byoung-Chul Lee and Ronald N. Zuckermann* Biological Nanostructures Facility, The Molecular Foundry, Lawrence Berkeley National Laboratory, 1 Cyclotron Rd., Berkeley, California 94720, United States bS Supporting Information ABSTRACT: For the last few decades, chemistry has played an important role in protein engineering by providing a variety of synthetic tools such as chemoselective side-chain modifications, chemical conjugation, incorporation of non-natural amino acids, and the development of protein-mimetic heteropolymers. Here we study protein backbone engineering in order to better understand the molecular mechanism of protein function and to introduce protease stable, non-natural residues into a protein structure. Using a combination of genetic engineering and chemical synthesis, we were able to introduce peptoid residues (N-substituted glycine residues) at defined positions into bovine pancreatic ribonuclease A. This results in a side-chain translocation from the Cα carbon to the neighboring backbone nitrogen atom. To generate these peptoid substitutions, we removed the N-terminal S-peptide of the protein by proteolysis and chemically conjugated synthetic peptide-peptoid hybrids to the new N-terminus. A triple peptoid mutant containing a catalytic  4 À1 À1 His12 peptoid mutation was active with a kcat/Km value of 1.0 10 M s . This kcat/Km value is only 10-fold lower than the control wild-type conjugate and comparable in magnitude to many other natural enzymes. The peptoid mutations increased the chain flexibility at the site of peptoid substitution and at its C-terminal neighboring residue. -

Discrete Arginine Topologies Guide Escape of Miniature Proteins from Early Endosomes to the Cytoplasm Jacob S
Yale University EliScholar – A Digital Platform for Scholarly Publishing at Yale Yale Medicine Thesis Digital Library School of Medicine 10-1-2012 Discrete arginine topologies guide escape of miniature proteins from early endosomes to the cytoplasm Jacob S. Appelbaum Yale University Follow this and additional works at: http://elischolar.library.yale.edu/ymtdl Part of the Medicine and Health Sciences Commons Recommended Citation Appelbaum, Jacob S., "Discrete arginine topologies guide escape of miniature proteins from early endosomes to the cytoplasm" (2012). Yale Medicine Thesis Digital Library. 3362. http://elischolar.library.yale.edu/ymtdl/3362 This Open Access Dissertation is brought to you for free and open access by the School of Medicine at EliScholar – A Digital Platform for Scholarly Publishing at Yale. It has been accepted for inclusion in Yale Medicine Thesis Digital Library by an authorized administrator of EliScholar – A Digital Platform for Scholarly Publishing at Yale. For more information, please contact [email protected]. Discrete arginine topologies guide escape of miniature proteins from early endosomes to the cytoplasm. A Dissertation Presented to the Faculty of the Graduate School of Yale University in Candidacy for the Degree of Doctor of Philosophy by Jacob S. Appelbaum Dissertation Director: Alanna Schepartz i October 2012 Abstract Discrete arginine topologies guide escape of miniature proteins from early endosomes to the cytoplasm. Jacob S. Appelbaum 2012 Polypeptides and peptide mimetics sample a wide chemical space with broad potential to modulate cellular function, but their application to cytoplasmic targets is limited be cause when added to cells their cytosolic concentration remains low. This limitation is due to a diffusion barrier (the plasma membrane) and absence of dedicated import ma chinery. -

Gifc-2018 Book of Abstracts
GIFC-2018 Giornate Italo-Francesi di Chimica Journées Franco-Italiennes de Chimie 16 – 18 April 2018 Grand Hotel Savoia Via Arsenale di Terra, 5 Genova BOOK OF ABSTRACTS ISBN: 978-88-94952-00-1 Gold Sponsors Silver Sponsors Patronages 2 3 Scientific Program 4 5 6 List of Posters (according to alphabetic order of presenting Authors) 1st session, Monday 16th April Num Cognome Nome Contatto Titolo Synthesis, purification and characterization of PO1 Aboudou Soioulata [email protected] antimicrobial peptides isolated from animal venoms [email protected] PO2 Ajmalghan Muthali Coverage dependent recombination mechanisms of hydrogen from niv-amu.fr tungsten surfaces via density functional theory Hybrid squaraine-silica nanoparticles as nir probes for biological PO3 Alberto Gabriele [email protected] applications: optimization of the photoemission performances Synthesis of doped metal oxide nanocrystals for solution-processed PO4 Alkarsifi Riva [email protected] interfacial layers in organic solar cells PO5 Anceschi Anastasia [email protected] Maltodextrins nanosponges as precursor for porous carbon materials [email protected] PO7 Arnodo Davide First racemic total synthesis of heliolactone .it [email protected]. PO8 Azzi Emanuele Synthesis of boronated analogue of curcumin as potential therapeutical it agents for alzheimer’s disease Study of Nanobiomaterials with Bio-based Antioxidants: Interaction of PO9 Barzan Giulia Sci piem Polyphenol Molecules with Hydroxyapatite and Silica Luigi Methotrexate loaded solid lipid nanoparticles: protein functionalization to PO10 Battaglia [email protected] Sebastiano improve brain biodistribution [email protected] PO11 Begni Federico On the adsorption of toluene on porous materials with different chemical m composition Synthesischaracterization of activated carbon from modified banana peels PO12 Ben Khalifa Eya [email protected] for hexavalent chromium adsorption [email protected] PO13 Benvenuti Martino The maturation of the co-dehydrogenase from thermococcus sp. -

Chiral Dualism As an Instrument of Hierarchical Structure Formation in Molecular Biology
S S symmetry Article Chiral Dualism as an Instrument of Hierarchical Structure Formation in Molecular Biology Vsevolod A. Tverdislov * and Ekaterina V. Malyshko * Faculty of Physics, Lomonosov Moscow State University, Leninskie gory 1-2, 119234 Moscow, Russia * Correspondence: [email protected] (V.A.T.); [email protected] (E.V.M.) Received: 6 March 2020; Accepted: 2 April 2020; Published: 8 April 2020 Abstract: The origin of chiral asymmetry in biology has attracted the attention of the research community throughout the years. In this paper we discuss the role of chirality and chirality sign alternation (L–D–L–D in proteins and D–L–D–L in DNA) in promoting self-organization in biology, starting at the level of single molecules and continuing to the level of supramolecular assemblies. In addition, we also discuss chiral assemblies in solutions of homochiral organic molecules. Sign-alternating chiral hierarchies created by proteins and nucleic acids are suggested to create the structural basis for the existence of selected mechanical degrees of freedom required for conformational dynamics in enzymes and macromolecular machines. Keywords: chirality; proteins; nucleic acids; hierarchy of structures; folding; molecular machines 1. Introduction The origin and potential role of chiral asymmetry in biology have attracted the attention of the research community throughout the years [1–18]. Although, at the macroscopic level, nonbiological structures with a homochiral molecular basis are not known, living organisms have been engaged -

Flexible Fluidic Actuators for Soft Robotic Applications
University of Wollongong Research Online University of Wollongong Thesis Collection 2017+ University of Wollongong Thesis Collections 2019 Flexible Fluidic Actuators for Soft Robotic Applications Weiping Hu University of Wollongong Follow this and additional works at: https://ro.uow.edu.au/theses1 University of Wollongong Copyright Warning You may print or download ONE copy of this document for the purpose of your own research or study. The University does not authorise you to copy, communicate or otherwise make available electronically to any other person any copyright material contained on this site. You are reminded of the following: This work is copyright. Apart from any use permitted under the Copyright Act 1968, no part of this work may be reproduced by any process, nor may any other exclusive right be exercised, without the permission of the author. Copyright owners are entitled to take legal action against persons who infringe their copyright. A reproduction of material that is protected by copyright may be a copyright infringement. A court may impose penalties and award damages in relation to offences and infringements relating to copyright material. Higher penalties may apply, and higher damages may be awarded, for offences and infringements involving the conversion of material into digital or electronic form. Unless otherwise indicated, the views expressed in this thesis are those of the author and do not necessarily represent the views of the University of Wollongong. Recommended Citation Hu, Weiping, Flexible Fluidic Actuators for Soft Robotic Applications, Doctor of Philosophy thesis, School of Mechanical, Materials, Mechatronic and Biomedical Engineering, University of Wollongong, 2019. https://ro.uow.edu.au/theses1/717 Research Online is the open access institutional repository for the University of Wollongong. -

Versatile Oligo(N-Substituted) Glycines: the Many Roles Of
WILEY-VCH K+V Fotosatz GmbH Nielsen Beerfelden 1. UK 19.11.2003 H.J. Maier 1 1 Versatile OligoN-Substituted) Glycines: The Many Roles of Peptoids in Drug Discovery James A. Patch, Kent Kirshenbaum, Shannon L. Seurynck, Ronald N. Zuckermann, and Annelise E. Barron 1.1 Introduction Despite their wide range of important bioactivities, polypeptides are generally poor drugs. Typically, they are rapidly degraded by proteases in vivo, and are fre- quently immunogenic. This fact has stimulated widespread efforts to develop pep- tide mimics for biomedical applications, a task that presents formidable chal- lenges in molecular design. Chemists seek efficient routes to peptidomimetic compounds with enhanced pharmacological properties, which retain the activities of their biological counterparts. Since peptides play myriad roles in living sys- tems, it is likely that no individual strategy will suffice. Indeed, a wide variety of different peptidomimetic oligomer scaffolds have been explored [1]. In order to ad- dress multiple design criteria for applications ranging from medicinal chemistry to materials science, researchers have worked to identify a non-natural chemical scaffold that recapitulates the desirable attributes of polypeptides. These include good solubility in aqueous solution, access to facile sequence-specific assembly of monomers containing chemically diverse side chains, and the capacity to form stable, biomimetic folded structures. Among the first reports of chemically diverse peptide mimics were those of %N- substituted) glycine oligomers %peptoids) [2]. Sequence-specific oligopeptoids have now been studied for over a decade, and have provided illustrative examples of both the potential of peptidomimetics and the obstacles faced in translating this potential into clinically useful compounds. -

Prion-Specific Peptoid Reagents Prionspezifische Peptoidreagentien Reactifs Peptoides Specifiques Des Prions
(19) TZZ_¥_5B_T (11) EP 1 931 695 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C07K 7/06 (2006.01) C07K 7/08 (2006.01) 10.04.2013 Bulletin 2013/15 A61K 38/08 (2006.01) A61K 38/10 (2006.01) A61K 31/16 (2006.01) G01N 33/68 (2006.01) (2006.01) (2006.01) (21) Application number: 06814415.3 G01N 33/50 A61P 25/28 C07K 14/47 (2006.01) (22) Date of filing: 08.09.2006 (86) International application number: PCT/US2006/035226 (87) International publication number: WO 2007/030804 (15.03.2007 Gazette 2007/11) (54) Prion-specific peptoid reagents Prionspezifische Peptoidreagentien Reactifs peptoides specifiques des prions (84) Designated Contracting States: (74) Representative: Marshall, Cameron John et al AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Carpmaels & Ransford HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI One Southampton Row SK TR London WC1B 5HA (GB) (30) Priority: 09.09.2005 US 715761 P 14.10.2005 US 726686 P (56) References cited: 13.01.2006 US 758934 P WO-A1-94/06451 WO-A2-02/052046 WO-A2-2005/016127 WO-A2-2005/060697 (43) Date of publication of application: US-A- 5 965 695 US-A1- 2003 040 468 18.06.2008 Bulletin 2008/25 • HAYNES, RUSSELL D. ET AL: "Comblike, (60) Divisional application: Monodisperse Polypeptoid Drag-Tags for DNA 11171585.0 / 2 383 281 Separations by End-Labeled Free-Solution Electrophoresis (ELFSE)" BIOCONJUGATE (73) Proprietor: Novartis AG CHEMISTRY, vol.